+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Architecture of the ESCPE-1 membrane coat | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | sorting nexins / PX domain / BAR domain / endosomes / retrograde transport / endocytic recycling / cargo recognition / protein coat / membrane recruitment / membrane deformation / membrane tubules. / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationretromer, tubulation complex / lamellipodium morphogenesis / epidermal growth factor catabolic process / pinocytosis / cytoplasmic side of early endosome membrane / leptin receptor binding / tubular endosome / macropinocytic cup / clathrin coat / early endosome to Golgi transport ...retromer, tubulation complex / lamellipodium morphogenesis / epidermal growth factor catabolic process / pinocytosis / cytoplasmic side of early endosome membrane / leptin receptor binding / tubular endosome / macropinocytic cup / clathrin coat / early endosome to Golgi transport / Retrograde transport at the Trans-Golgi-Network / response to tetrachloromethane / insulin-like growth factor receptor activity / retromer complex binding / insulin-like growth factor binding / phosphatidylinositol-5-phosphate binding / transferrin receptor binding / retromer complex / insulin-like growth factor II binding / trans-Golgi network transport vesicle / host-mediated activation of viral process / retinoic acid binding / phosphatidylinositol-4-phosphate binding / retrograde transport, endosome to Golgi / phosphatidylinositol-3,5-bisphosphate binding / epidermal growth factor receptor binding / lysosomal transport / Golgi Associated Vesicle Biogenesis / nuclear envelope lumen / D-mannose binding / brush border / dynactin binding / animal organ regeneration / endocytic vesicle / G-protein alpha-subunit binding / regulation of macroautophagy / positive regulation of insulin receptor signaling pathway / response to retinoic acid / D1 dopamine receptor binding / phagocytic cup / transport vesicle / negative regulation of blood pressure / ruffle / phosphatidylinositol binding / receptor-mediated endocytosis / secretory granule membrane / trans-Golgi network membrane / post-embryonic development / insulin receptor binding / intracellular protein transport / phosphoprotein binding / trans-Golgi network / clathrin-coated endocytic vesicle membrane / liver development / receptor internalization / cytoplasmic side of plasma membrane / positive regulation of protein catabolic process / late endosome / Cargo recognition for clathrin-mediated endocytosis / signaling receptor activity / lamellipodium / Clathrin-mediated endocytosis / early endosome membrane / spermatogenesis / vesicle / early endosome / lysosome / endosome / endosome membrane / positive regulation of apoptotic process / cadherin binding / G protein-coupled receptor signaling pathway / protein heterodimerization activity / Golgi membrane / focal adhesion / intracellular membrane-bounded organelle / Neutrophil degranulation / positive regulation of DNA-templated transcription / perinuclear region of cytoplasm / enzyme binding / cell surface / Golgi apparatus / signal transduction / protein homodimerization activity / protein-containing complex / extracellular exosome / identical protein binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
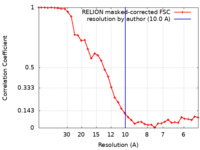
| Method | subtomogram averaging / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Lopez-Robles C / Scaramuzza S / Astorga-Simon E / Ishida M / Williamsom CD / Banos-Mateos S / Gil-Carton D / Romero M / Vidaurrazaga A / Fernandez-Recio J ...Lopez-Robles C / Scaramuzza S / Astorga-Simon E / Ishida M / Williamsom CD / Banos-Mateos S / Gil-Carton D / Romero M / Vidaurrazaga A / Fernandez-Recio J / Rojas AL / Bonifacino JS / Castano-Diez D / Hierro A | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Architecture of the ESCPE-1 membrane coat. Authors: Carlos Lopez-Robles / Stefano Scaramuzza / Elsa N Astorga-Simon / Morié Ishida / Chad D Williamson / Soledad Baños-Mateos / David Gil-Carton / Miguel Romero-Durana / Ander Vidaurrazaga / ...Authors: Carlos Lopez-Robles / Stefano Scaramuzza / Elsa N Astorga-Simon / Morié Ishida / Chad D Williamson / Soledad Baños-Mateos / David Gil-Carton / Miguel Romero-Durana / Ander Vidaurrazaga / Juan Fernandez-Recio / Adriana L Rojas / Juan S Bonifacino / Daniel Castaño-Díez / Aitor Hierro /    Abstract: Recycling of membrane proteins enables the reuse of receptors, ion channels and transporters. A key component of the recycling machinery is the endosomal sorting complex for promoting exit 1 (ESCPE-1) ...Recycling of membrane proteins enables the reuse of receptors, ion channels and transporters. A key component of the recycling machinery is the endosomal sorting complex for promoting exit 1 (ESCPE-1), which rescues transmembrane proteins from the endolysosomal pathway for transport to the trans-Golgi network and the plasma membrane. This rescue entails the formation of recycling tubules through ESCPE-1 recruitment, cargo capture, coat assembly and membrane sculpting by mechanisms that remain largely unknown. Herein, we show that ESCPE-1 has a single-layer coat organization and suggest how synergistic interactions between ESCPE-1 protomers, phosphoinositides and cargo molecules result in a global arrangement of amphipathic helices to drive tubule formation. Our results thus define a key process of tubule-based endosomal sorting. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15413.map.gz emd_15413.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15413-v30.xml emd-15413-v30.xml emd-15413.xml emd-15413.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15413_fsc.xml emd_15413_fsc.xml | 3.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15413.png emd_15413.png | 161.3 KB | ||
| Filedesc metadata |  emd-15413.cif.gz emd-15413.cif.gz | 6.5 KB | ||
| Others |  emd_15413_half_map_1.map.gz emd_15413_half_map_1.map.gz emd_15413_half_map_2.map.gz emd_15413_half_map_2.map.gz | 3.1 MB 3.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15413 http://ftp.pdbj.org/pub/emdb/structures/EMD-15413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15413 | HTTPS FTP |
-Related structure data
| Related structure data |  8afzMC  8a1gC  8abqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15413.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15413.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.73 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15413_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15413_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Endosomal Sorting Complex for Promoting Exit 1 (ESCPE-1)
| Entire | Name: Endosomal Sorting Complex for Promoting Exit 1 (ESCPE-1) |
|---|---|
| Components |
|
-Supramolecule #1: Endosomal Sorting Complex for Promoting Exit 1 (ESCPE-1)
| Supramolecule | Name: Endosomal Sorting Complex for Promoting Exit 1 (ESCPE-1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: Sorting nexin-1
| Macromolecule | Name: Sorting nexin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.14434 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASGGGGCSA SERLPPPFPG LEPESEGAAG GSEPEAGDSD TEGEDIFTGA AVVSKHQSPK ITTSLLPINN GSKENGIHEE QDQEPQDLF ADATVELSLD STQNNQKKVL AKTLISLPPQ EATNSSKPQP TYEELEEEEQ EDQFDLTVGI TDPEKIGDGM N AYVAYKVT ...String: MASGGGGCSA SERLPPPFPG LEPESEGAAG GSEPEAGDSD TEGEDIFTGA AVVSKHQSPK ITTSLLPINN GSKENGIHEE QDQEPQDLF ADATVELSLD STQNNQKKVL AKTLISLPPQ EATNSSKPQP TYEELEEEEQ EDQFDLTVGI TDPEKIGDGM N AYVAYKVT TQTSLPLFRS KQFAVKRRFS DFLGLYEKLS EKHSQNGFIV PPPPEKSLIG MTKVKVGKED SSSAEFLEKR RA ALERYLQ RIVNHPTMLQ DPDVREFLEK EELPRAVGTQ TLSGAGLLKM FNKATDAVSK MTIKMNESDI WFEEKLQEVE CEE QRLRKL HAVVETLVNH RKELALNTAQ FAKSLAMLGS SEDNTALSRA LSQLAEVEEK IEQLHQEQAN NDFFLLAELL SDYI RLLAI VRAAFDQRMK TWQRWQDAQA TLQKKREAEA RLLWANKPDK LQQAKDEILE WESRVTQYER DFERISTVVR KEVIR FEKE KSKDFKNHVI KYLETLLYSQ QQLAKYWEAF LPEAKAIS UniProtKB: Sorting nexin-1 |
-Macromolecule #2: Sorting nexin-5
| Macromolecule | Name: Sorting nexin-5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.891504 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAVPELLQQ QEEDRSKLRS VSVDLNVDPS LQIDIPDALS ERDKVKFTVH TKTTLPTFQS PEFSVTRQHE DFVWLHDTLI ETTDYAGLI IPPAPTKPDF DGPREKMQKL GEGEGSMTKE EFAKMKQELE AEYLAVFKKT VSSHEVFLQR LSSHPVLSKD R NFHVFLEY ...String: MAAVPELLQQ QEEDRSKLRS VSVDLNVDPS LQIDIPDALS ERDKVKFTVH TKTTLPTFQS PEFSVTRQHE DFVWLHDTLI ETTDYAGLI IPPAPTKPDF DGPREKMQKL GEGEGSMTKE EFAKMKQELE AEYLAVFKKT VSSHEVFLQR LSSHPVLSKD R NFHVFLEY DQDLSVRRKN TKEMFGGFFK SVVKSADEVL FTGVKEVDDF FEQEKNFLIN YYNRIKDSCV KADKMTRSHK NV ADDYIHT AACLHSLALE EPTVIKKYLL KVAELFEKLR KVEGRVSSDE DLKLTELLRY YMLNIEAAKD LLYRRTKALI DYE NSNKAL DKARLKSKDV KLAEAHQQEC CQKFEQLSES AKEELINFKR KRVAAFRKNL IEMSELEIKH ARNNVSLLQS CIDL FKNN UniProtKB: Sorting nexin-5 |
-Macromolecule #3: Cation-independent mannose-6-phosphate receptor
| Macromolecule | Name: Cation-independent mannose-6-phosphate receptor / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.766033 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNVSYKYSKV NKEEETDENE TEWLMEEIQL P UniProtKB: Cation-independent mannose-6-phosphate receptor |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 282 K / Instrument: FEI VITROBOT MARK II / Details: Incubation time 30s Blotting time 2s. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average electron dose: 2.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)