[English] 日本語
 Yorodumi
Yorodumi- EMDB-15389: 3 A CRYO-EM STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS FERRITIN FROM... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3 A CRYO-EM STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS FERRITIN FROM TIMEPIX3 detector | |||||||||
 Map data Map data | The map is sharpened by Locspiral. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | IRON STORAGE / FERROXIDASE / BACTERIAL FERRITIN / OCTAHEDRAL SYMMETRY. / METAL TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationMtb iron assimilation by chelation / response to nitrosative stress / encapsulin nanocompartment / iron ion sequestering activity / ferroxidase / ferroxidase activity / intracellular sequestering of iron ion / ferric iron binding / peptidoglycan-based cell wall / ferrous iron binding ...Mtb iron assimilation by chelation / response to nitrosative stress / encapsulin nanocompartment / iron ion sequestering activity / ferroxidase / ferroxidase activity / intracellular sequestering of iron ion / ferric iron binding / peptidoglycan-based cell wall / ferrous iron binding / iron ion transport / response to hypoxia / extracellular region / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | |||||||||
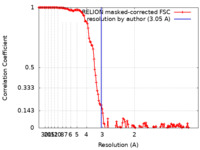
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Zhang Y / van Schayck JP / Knoops K / Peters PJ / Ravelli RBG | |||||||||
| Funding support |  Netherlands, European Union, 2 items Netherlands, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2021 Journal: Acta Crystallogr D Struct Biol / Year: 2021Title: Mycobacterium tuberculosis ferritin: a suitable workhorse protein for cryo-EM development. Authors: Abril Gijsbers / Yue Zhang / Ye Gao / Peter J Peters / Raimond B G Ravelli /  Abstract: The use of cryo-EM continues to expand worldwide and calls for good-quality standard proteins with simple protocols for their production. Here, a straightforward expression and purification protocol ...The use of cryo-EM continues to expand worldwide and calls for good-quality standard proteins with simple protocols for their production. Here, a straightforward expression and purification protocol is presented that provides an apoferritin, bacterioferritin B (BfrB), from Mycobacterium tuberculosis with high yield and purity. A 2.12 Å resolution cryo-EM structure of BfrB is reported, showing the typical cage-like oligomer constituting of 24 monomers related by 432 symmetry. However, it also contains a unique C-terminal extension (164-181), which loops into the cage region of the shell and provides extra stability to the protein. Part of this region was ambiguous in previous crystal structures but could be built within the cryo-EM map. These findings and this protocol could serve the growing cryo-EM community in characterizing and pushing the limits of their electron microscopes and workflows. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15389.map.gz emd_15389.map.gz | 37 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15389-v30.xml emd-15389-v30.xml emd-15389.xml emd-15389.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15389_fsc.xml emd_15389_fsc.xml | 13.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_15389.png emd_15389.png | 81.1 KB | ||
| Masks |  emd_15389_msk_1.map emd_15389_msk_1.map | 229.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15389.cif.gz emd-15389.cif.gz | 6.2 KB | ||
| Others |  emd_15389_additional_1.map.gz emd_15389_additional_1.map.gz emd_15389_half_map_1.map.gz emd_15389_half_map_1.map.gz emd_15389_half_map_2.map.gz emd_15389_half_map_2.map.gz | 214.8 MB 175.9 MB 175.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15389 http://ftp.pdbj.org/pub/emdb/structures/EMD-15389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15389 | HTTPS FTP |
-Validation report
| Summary document |  emd_15389_validation.pdf.gz emd_15389_validation.pdf.gz | 916.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15389_full_validation.pdf.gz emd_15389_full_validation.pdf.gz | 916.5 KB | Display | |
| Data in XML |  emd_15389_validation.xml.gz emd_15389_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  emd_15389_validation.cif.gz emd_15389_validation.cif.gz | 28.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15389 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15389 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15389 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15389 | HTTPS FTP |
-Related structure data
| Related structure data |  8aeyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15389.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15389.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map is sharpened by Locspiral. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.6 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15389_msk_1.map emd_15389_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: The map is sharpened by Relion postprocessing.
| File | emd_15389_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map is sharpened by Relion postprocessing. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15389_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15389_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MYCOBACTERIUM TUBERCULOSIS FERRITIN
| Entire | Name: MYCOBACTERIUM TUBERCULOSIS FERRITIN |
|---|---|
| Components |
|
-Supramolecule #1: MYCOBACTERIUM TUBERCULOSIS FERRITIN
| Supramolecule | Name: MYCOBACTERIUM TUBERCULOSIS FERRITIN / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: MYCOBACTERIUM TUBERCULOSIS FERRITIN |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
-Macromolecule #1: Ferritin BfrB
| Macromolecule | Name: Ferritin BfrB / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO / EC number: ferroxidase |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 20.463936 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTEYEGPKTK FHALMQEQIH NEFTAAQQYV AIAVYFDSED LPQLAKHFYS QAVEERNHAM MLVQHLLDRD LRVEIPGVDT VRNQFDRPR EALALALDQE RTVTDQVGRL TAVARDEGDF LGEQFMQWFL QEQIEEVALM ATLVRVADRA GANLFELENF V AREVDVAP AASGAPHAAG GRL UniProtKB: Bacterioferritin BfrB |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 240 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 40 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Software | Name: SerialEM (ver. 4.0) |
| Details | BASIC DIRECT ALIGNMENTS WERE DONE AS WELL AS ASTIGMATISM AND COMA ALIGNMENT USING AUTOCTF |
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 512 pixel / Digitization - Dimensions - Height: 512 pixel / Number real images: 2977 / Average exposure time: 1.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing #1
Image processing #1
+ Image processing #2
Image processing #2
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 79.7 / Target criteria: CORRELATION COEFFICIENT |
| Output model |  PDB-8aey: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)