[English] 日本語
 Yorodumi
Yorodumi- EMDB-15241: Mycobacterium tuberculosis ClpC1 hexamer structure bound to the n... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin | ||||||||||||
 Map data Map data | Sharpened map of Mycobacterium tuberculosis ClpC1 cyclomarin | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein folding chaperone / peptidoglycan-based cell wall / cellular response to heat / protein homodimerization activity / ATP hydrolysis activity / ATP binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
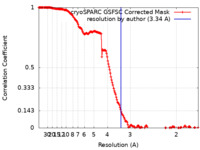
| Method | single particle reconstruction / cryo EM / Resolution: 3.34 Å | ||||||||||||
 Authors Authors | Felix J / Fraga H / Gragera M / Bueno T / Weinhaeupl K | ||||||||||||
| Funding support | European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action. Authors: Katharina Weinhäupl / Marcos Gragera / M Teresa Bueno-Carrasco / Rocío Arranz / Olga Krandor / Tatos Akopian / Raquel Soares / Eric Rubin / Jan Felix / Hugo Fraga /     Abstract: The unfoldase ClpC1 is one of the most exciting drug targets against tuberculosis. This AAA+ unfoldase works in cooperation with the ClpP1P2 protease and is the target of at least four natural ...The unfoldase ClpC1 is one of the most exciting drug targets against tuberculosis. This AAA+ unfoldase works in cooperation with the ClpP1P2 protease and is the target of at least four natural product antibiotics: cyclomarin, ecumicin, lassomycin, and rufomycin. Although these molecules are promising starting points for drug development, their mechanisms of action remain largely unknown. Taking advantage of a middle domain mutant, we determined the first structure of Mycobacterium tuberculosis ClpC1 in its apo, cyclomarin-, and ecumicin-bound states via cryo-EM. The obtained structure displays features observed in other members of the AAA+ family and provides a map for further drug development. While the apo and cyclomarin-bound structures are indistinguishable and have N-terminal domains that are invisible in their respective EM maps, around half of the ecumicin-bound ClpC1 particles display three of their six N-terminal domains in an extended conformation. Our structural observations suggest a mechanism where ecumicin functions by mimicking substrate binding, leading to ATPase activation and changes in protein degradation profile. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15241.map.gz emd_15241.map.gz | 450.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15241-v30.xml emd-15241-v30.xml emd-15241.xml emd-15241.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15241_fsc.xml emd_15241_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_15241.png emd_15241.png | 101.1 KB | ||
| Masks |  emd_15241_msk_1.map emd_15241_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Others |  emd_15241_additional_1.map.gz emd_15241_additional_1.map.gz emd_15241_half_map_1.map.gz emd_15241_half_map_1.map.gz emd_15241_half_map_2.map.gz emd_15241_half_map_2.map.gz | 421.7 MB 442.4 MB 442.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15241 http://ftp.pdbj.org/pub/emdb/structures/EMD-15241 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15241 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15241 | HTTPS FTP |
-Validation report
| Summary document |  emd_15241_validation.pdf.gz emd_15241_validation.pdf.gz | 853.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15241_full_validation.pdf.gz emd_15241_full_validation.pdf.gz | 853.1 KB | Display | |
| Data in XML |  emd_15241_validation.xml.gz emd_15241_validation.xml.gz | 25.9 KB | Display | |
| Data in CIF |  emd_15241_validation.cif.gz emd_15241_validation.cif.gz | 34.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15241 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15241 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15241 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15241 | HTTPS FTP |
-Related structure data
| Related structure data |  8a8vMC  8a8uC  8a8wC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15241.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15241.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of Mycobacterium tuberculosis ClpC1 cyclomarin | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||
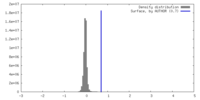
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15241_msk_1.map emd_15241_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
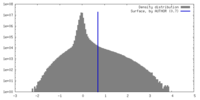
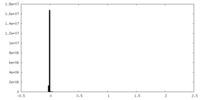
| Density Histograms |
-Additional map: DeepEMhancer sharpened map of Mycobacterium tuberculosis ClpC1 cyclomarin...
| File | emd_15241_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map of Mycobacterium tuberculosis ClpC1 cyclomarin | ||||||||||||
| Projections & Slices |
| ||||||||||||
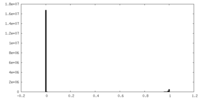
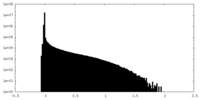
| Density Histograms |
-Half map: #2
| File | emd_15241_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
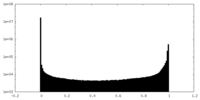
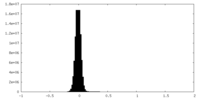
| Density Histograms |
-Half map: #1
| File | emd_15241_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
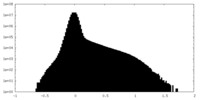
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterium tuberculosis ClpC1 in complex with the natural prod...
| Entire | Name: Mycobacterium tuberculosis ClpC1 in complex with the natural product antibiotic Cyclomarin |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium tuberculosis ClpC1 in complex with the natural prod...
| Supramolecule | Name: Mycobacterium tuberculosis ClpC1 in complex with the natural product antibiotic Cyclomarin type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 567 KDa |
-Macromolecule #1: ATP-dependent Clp protease ATP-binding subunit ClpC1
| Macromolecule | Name: ATP-dependent Clp protease ATP-binding subunit ClpC1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: Hydrolases; Acting on peptide bonds (peptidases) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 94.758695 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFERFTDRAR RVVVLAQEEA RMLNHNYIGT EHILLGLIHE GEGVAAKSLE SLGISLEGVR SQVEEIIGQG QQAPSGHIPF TPRAKKVLE LSLREALQLG HNYIGTEHIL LGLIREGEGV AAQVLVKLGA ELTRVRQQVI QLLSGYQGKE AAEAGTGGRG G ESGSPSTS ...String: MFERFTDRAR RVVVLAQEEA RMLNHNYIGT EHILLGLIHE GEGVAAKSLE SLGISLEGVR SQVEEIIGQG QQAPSGHIPF TPRAKKVLE LSLREALQLG HNYIGTEHIL LGLIREGEGV AAQVLVKLGA ELTRVRQQVI QLLSGYQGKE AAEAGTGGRG G ESGSPSTS LVLDQFGRNL TAAAMEGKLD PVIGREKEIE RVMQVLSRRT KNNPVLIGEP GVGKTAVVEG LAQAIVHGEV PE TLKDKQL YTLDLGSLVA GSRYRGDFEE RLKKVLKEIN TRGDIILFID ELHTLVGAGA AEGAIDAASI LKPKLARGEL QTI GATTLD EYRKYIEKDA ALERRFQPVQ VGEPTVEHTI EILKGLRDRY EAHHRVSITD AAMVAAATLA DRYINDRFLP DKAI DLIDE AGARMRIRRM TAPPDLREFD EKIAEARREK ESAIDAQDFE KAASLRDREK TLVAQRAERE KQWRSGDLDV VAEVD DEQI AEVLGNWTGI PVFKLTEAET TRLLRMEEEL HKRIIGQEDA VKAVSKAIRR TRAGLKDPKR PSGSFIFAGP SGVGKT ELS KALANFLFGD DDALIQIDMG EFHDRFTASR LFGAPPGYVG YEEGGQLTEK VRRKPFSVVL FDEIEKAHQE IYNSLLQ VL EDGRLTDGQG RTVDFKNTVL IFTSNLGTSD ISKPVGLGFS KGGGENDYER MKQKVNDELK KHFRPEFLNR IDDIIVFH Q LTREEIIRMV DLMISRVAGQ LKSKDMALVL TDAAKALLAK RGFDPVLGAR PLRRTIQREI EDQLSEKILF EEVGPGQVV TVDVDNWDGE GPGEDAVFTF TGTRKPPAEP DLAKAGAHSA GGPEPAARLE HHHHHH |
-Macromolecule #2: Bound polypeptide
| Macromolecule | Name: Bound polypeptide / type: protein_or_peptide / ID: 2 / Details: The actual sequence of the polypeptide is unknown. / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.975426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 10 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: Hepes pH 7.4 50 mM, NaCl 100 mM, 10 mM MgCl2, ATP 1mM |
| Vitrification | Cryogen name: ETHANE |
| Details | 10 microM ClpC1 with 30 microM Cyclomarin |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average exposure time: 30.0 sec. / Average electron dose: 36.9 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 3.1 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)