+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plastocyanin bound to PSI of Chlamydomonas reinhardtii | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Photosynthesis / green algae / dimeric PSI | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast thylakoid membrane / electron transfer activity / copper ion binding Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
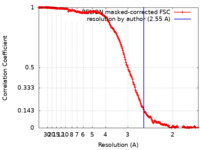
| Method | single particle reconstruction / cryo EM / Resolution: 2.55 Å | |||||||||||||||
 Authors Authors | Naschberger A / Amunts A | |||||||||||||||
| Funding support |  Sweden, Sweden,  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2022 Journal: Nat Plants / Year: 2022Title: Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex. Authors: Andreas Naschberger / Laura Mosebach / Victor Tobiasson / Sebastian Kuhlgert / Martin Scholz / Annemarie Perez-Boerema / Thi Thu Hoai Ho / André Vidal-Meireles / Yuichiro Takahashi / ...Authors: Andreas Naschberger / Laura Mosebach / Victor Tobiasson / Sebastian Kuhlgert / Martin Scholz / Annemarie Perez-Boerema / Thi Thu Hoai Ho / André Vidal-Meireles / Yuichiro Takahashi / Michael Hippler / Alexey Amunts /     Abstract: Photosystem I (PSI) enables photo-electron transfer and regulates photosynthesis in the bioenergetic membranes of cyanobacteria and chloroplasts. Being a multi-subunit complex, its macromolecular ...Photosystem I (PSI) enables photo-electron transfer and regulates photosynthesis in the bioenergetic membranes of cyanobacteria and chloroplasts. Being a multi-subunit complex, its macromolecular organization affects the dynamics of photosynthetic membranes. Here we reveal a chloroplast PSI from the green alga Chlamydomonas reinhardtii that is organized as a homodimer, comprising 40 protein subunits with 118 transmembrane helices that provide scaffold for 568 pigments. Cryogenic electron microscopy identified that the absence of PsaH and Lhca2 gives rise to a head-to-head relative orientation of the PSI-light-harvesting complex I monomers in a way that is essentially different from the oligomer formation in cyanobacteria. The light-harvesting protein Lhca9 is the key element for mediating this dimerization. The interface between the monomers is lacking PsaH and thus partially overlaps with the surface area that would bind one of the light-harvesting complex II complexes in state transitions. We also define the most accurate available PSI-light-harvesting complex I model at 2.3 Å resolution, including a flexibly bound electron donor plastocyanin, and assign correct identities and orientations to all the pigments, as well as 621 water molecules that affect energy transfer pathways. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14872.map.gz emd_14872.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14872-v30.xml emd-14872-v30.xml emd-14872.xml emd-14872.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14872_fsc.xml emd_14872_fsc.xml | 17.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14872.png emd_14872.png | 67.6 KB | ||
| Filedesc metadata |  emd-14872.cif.gz emd-14872.cif.gz | 5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14872 http://ftp.pdbj.org/pub/emdb/structures/EMD-14872 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14872 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14872 | HTTPS FTP |
-Validation report
| Summary document |  emd_14872_validation.pdf.gz emd_14872_validation.pdf.gz | 337.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14872_full_validation.pdf.gz emd_14872_full_validation.pdf.gz | 337.3 KB | Display | |
| Data in XML |  emd_14872_validation.xml.gz emd_14872_validation.xml.gz | 16.6 KB | Display | |
| Data in CIF |  emd_14872_validation.cif.gz emd_14872_validation.cif.gz | 22.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14872 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14872 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14872 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14872 | HTTPS FTP |
-Related structure data
| Related structure data |  7zqeMC  7zq9C  7zqcC  7zqdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14872.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14872.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Photosystem I dimer of Chlamydomonas reinhardtii
| Entire | Name: Photosystem I dimer of Chlamydomonas reinhardtii |
|---|---|
| Components |
|
-Supramolecule #1: Photosystem I dimer of Chlamydomonas reinhardtii
| Supramolecule | Name: Photosystem I dimer of Chlamydomonas reinhardtii / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 700 KDa |
-Macromolecule #1: Plastocyanin, chloroplastic
| Macromolecule | Name: Plastocyanin, chloroplastic / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.889784 KDa |
| Sequence | String: MKATLRAPAS RASAVRPVAS LKAAAQRVAS VAGVSVASLA LTLAAHADAT VKLGADSGAL EFVPKTLTIK SGETVNFVNN AGFPHNIVF DEDAIPSGVN ADAISRDDYL NAPGETYSVK LTAAGEYGYY CEPHQGAGMV GKIIVQ UniProtKB: Plastocyanin, chloroplastic |
-Macromolecule #2: COPPER (II) ION
| Macromolecule | Name: COPPER (II) ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: CU |
|---|---|
| Molecular weight | Theoretical: 63.546 Da |
| Chemical component information |  ChemComp-CU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 45.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)