+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of POLRMT mutant. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondria / DNA dependent RNA polymerase. / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / Strand-asynchronous mitochondrial DNA replication / mitochondrial DNA replication / mitochondrial transcription / mitochondrial nucleoid / Transcriptional activation of mitochondrial biogenesis / DNA-directed RNA polymerase ...Mitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / Strand-asynchronous mitochondrial DNA replication / mitochondrial DNA replication / mitochondrial transcription / mitochondrial nucleoid / Transcriptional activation of mitochondrial biogenesis / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / 3'-5'-RNA exonuclease activity / sequence-specific DNA binding / mitochondrial matrix / protein-containing complex / mitochondrion / RNA binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Das H / Hallberg BM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization. Authors: Xuefeng Zhu / Xie Xie / Hrishikesh Das / Benedict G Tan / Yonghong Shi / Ali Al-Behadili / Bradley Peter / Elisa Motori / Sebastian Valenzuela / Viktor Posse / Claes M Gustafsson / B Martin ...Authors: Xuefeng Zhu / Xie Xie / Hrishikesh Das / Benedict G Tan / Yonghong Shi / Ali Al-Behadili / Bradley Peter / Elisa Motori / Sebastian Valenzuela / Viktor Posse / Claes M Gustafsson / B Martin Hällberg / Maria Falkenberg /   Abstract: The mitochondrial genome encodes 13 components of the oxidative phosphorylation system, and altered mitochondrial transcription drives various human pathologies. A polyadenylated, non-coding RNA ...The mitochondrial genome encodes 13 components of the oxidative phosphorylation system, and altered mitochondrial transcription drives various human pathologies. A polyadenylated, non-coding RNA molecule known as 7S RNA is transcribed from a region immediately downstream of the light strand promoter in mammalian cells, and its levels change rapidly in response to physiological conditions. Here, we report that 7S RNA has a regulatory function, as it controls levels of mitochondrial transcription both in vitro and in cultured human cells. Using cryo-EM, we show that POLRMT dimerization is induced by interactions with 7S RNA. The resulting POLRMT dimer interface sequesters domains necessary for promoter recognition and unwinding, thereby preventing transcription initiation. We propose that the non-coding 7S RNA molecule is a component of a negative feedback loop that regulates mitochondrial transcription in mammalian cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14619.map.gz emd_14619.map.gz | 87.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14619-v30.xml emd-14619-v30.xml emd-14619.xml emd-14619.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14619.png emd_14619.png | 90.6 KB | ||
| Filedesc metadata |  emd-14619.cif.gz emd-14619.cif.gz | 6.3 KB | ||
| Others |  emd_14619_half_map_1.map.gz emd_14619_half_map_1.map.gz emd_14619_half_map_2.map.gz emd_14619_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14619 http://ftp.pdbj.org/pub/emdb/structures/EMD-14619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14619 | HTTPS FTP |
-Related structure data
| Related structure data |  7zc4MC  7pzpC  7pzrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14619.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14619.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_14619_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
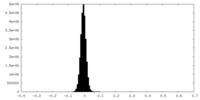
| Density Histograms |
-Half map: #2
| File | emd_14619_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
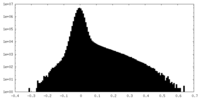
| Density Histograms |
- Sample components
Sample components
-Entire : POLRMT
| Entire | Name: POLRMT |
|---|---|
| Components |
|
-Supramolecule #1: POLRMT
| Supramolecule | Name: POLRMT / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all / Details: DNA-directed RNA polymerase, mitochondrial |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137 KDa |
-Macromolecule #1: DNA-directed RNA polymerase, mitochondrial
| Macromolecule | Name: DNA-directed RNA polymerase, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 134.327703 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QSSSASPQEQ DQDRRKDWGH VELLEVLQAR VRQLQAESVS EVVVNRVDVA RLPECGSGDG SLQPPRKVQM GAKDATPVPC GRWAKILEK DKRTQQMRMQ RLKAKLQMPF QSGEFKALTR RLQVEPRLLS KQMAGCLEDC TRQAPESPWE EQLARLLQEA P GKLSLDVE ...String: QSSSASPQEQ DQDRRKDWGH VELLEVLQAR VRQLQAESVS EVVVNRVDVA RLPECGSGDG SLQPPRKVQM GAKDATPVPC GRWAKILEK DKRTQQMRMQ RLKAKLQMPF QSGEFKALTR RLQVEPRLLS KQMAGCLEDC TRQAPESPWE EQLARLLQEA P GKLSLDVE QAPSGQHSQA QLSGQQQRLL AFFKCCLLTD QLPLAHHLLV VHHGQAQKRA LLTLDMYNAV MLGWARQGAF KE LVYVLFM VKDAGLTPDL LSYAAALQCM GRQDQDAGTI ERCLEQMSQE GLKLQALFTA VLLSEEDRAT VLKAVHKVKP TFS LPPQLP PPVNTSKLLR DVYAKDGRVS YPKLHLPLKT LQCLFEKQLH MELASRVCVV SVEKPTLPSK EVKHARATLA TLRD QWEKA LCRALRETKN RLEREVYEGR FSLYPFLCLL DEREVVRMLL QVLQALPAQG ESFTTLAREL SARTFSRHVV QRQRV SGQV QALQNHYRKY LCLLASDAEV PEPCLPRQYW EELGAPEALR EQPWPLPVQM ELGKLLAEML VQATQMPCSL DKPHRS SRL VPVLYHVYSF RNVQQIGILK PHPAYVQLLE KAAEPTLTFE AVDVPMLCPP LPWTSPHSGA FLLSPTKLMR TVEGATQ HQ ELLETCPPTA LHGALDALTQ LGNCAWRVNG RVLDLVLQLF QAKGCPQLGV PAPPSEAPQP PEAHLPHSAA PARKAELR R ELAHCQKVAR EMHSLRAEAL YRLSLAQHLR DRVFWLPHNM DFRGRTYPCP PHFNHLGSDV ARALLEFAQG RPLGPHGLD WLKIHLVNLT GLKKREPLRK RLAFAEEVMD DILDSADQPL TGRKWWMGAE EPWQTLACCM EVANAVRASD PAAYVSHLPV HQDGSCNGL QHYAALGRDS VGAASVNLEP SDVPQDVYSG VAAQVEVFRR QDAQRGMRVA QVLEGFITRK VVKQTVMTVV Y GVTRYGGR LQIEKRLREL SDFPQEFVWE ASHYLVAQVF ASLQEMFSGT RAIQHWLTES ARLISHMGSV VEWVTPLGVP VI QPYRLDS KVKQIGGGIQ SITYTHNGDI SRAPNTAKQA NGFPPNFIHS LDSSHMMLTA LHCYRKGLTF VSVHDCYWTH AAD VSVMNQ VCREQFVRLH SEPILQDLSR FLVKRFCSEP QKILEASQLK ETLQAVPKPG AFDLEQVKRS TYFFS UniProtKB: DNA-directed RNA polymerase, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6583 / Average exposure time: 1.5 sec. / Average electron dose: 48.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 20.0 µm / Calibrated defocus min: 0.3 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)