+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The molybdenum storage protein loaded with tungstate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Metal storage protein / polytungstate / Keggin ion / EM / METAL BINDING PROTEIN | |||||||||
| Function / homology | Molybdenum storage protein subunit alpha/beta / nutrient reservoir activity / molybdenum ion binding / Aspartate/glutamate/uridylate kinase / Acetylglutamate kinase-like superfamily / Amino acid kinase family / cytoplasm / Molybdenum storage protein subunit beta / Molybdenum storage protein subunit alpha Function and homology information Function and homology information | |||||||||
| Biological species |  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) | |||||||||
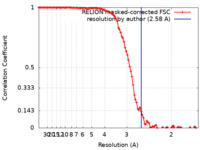
| Method | single particle reconstruction / cryo EM / Resolution: 2.58 Å | |||||||||
 Authors Authors | Ermler U / Aziz I / Kaltwasser S / Kayastha K / Vonck J | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: J Inorg Biochem / Year: 2022 Journal: J Inorg Biochem / Year: 2022Title: The molybdenum storage protein forms and deposits distinct polynuclear tungsten oxygen aggregates. Authors: Iram Aziz / Susann Kaltwasser / Kanwal Kayastha / Radhika Khera / Janet Vonck / Ulrich Ermler /  Abstract: Some N-fixing bacteria store Mo to maintain the formation of the vital FeMo-cofactor dependent nitrogenase under Mo depleting conditions. The Mo storage protein (MoSto), developed for this purpose, ...Some N-fixing bacteria store Mo to maintain the formation of the vital FeMo-cofactor dependent nitrogenase under Mo depleting conditions. The Mo storage protein (MoSto), developed for this purpose, has the unique capability to compactly deposit molybdate as polyoxometalate (POM) clusters in a (αβ) hexameric cage; the same occurs with the physicochemically related tungstate. To explore the structural diversity of W-based POM clusters, MoSto loaded under different conditions with tungstate and two site-specifically modified MoSto variants were structurally characterized by X-ray crystallography or single-particle cryo-EM. The MoSto cage contains five major locations for POM clusters occupied among others by heptanuclear, Keggin ion and even Dawson-like species also found in bulk solvent under defined conditions. We found both lacunary derivatives of these archetypical POM clusters with missing WO units at positions exposed to bulk solvent and expanded derivatives with additional WO units next to protecting polypeptide segments or other POM clusters. The cryo-EM map, unexpectedly, reveals a POM cluster in the cage center anchored to the wall by a WO linker. Interestingly, distinct POM cluster structures can originate from identical, highly occupied core fragments of three to seven WO units that partly correspond to those found in MoSto loaded with molybdate. These core fragments are firmly bound to the complementary protein template in contrast to the more variable, less occupied residual parts of the visible POM clusters. Due to their higher stability, W-based POM clusters are, on average, larger and more diverse than their Mo-based counterparts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14522.map.gz emd_14522.map.gz | 64.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14522-v30.xml emd-14522-v30.xml emd-14522.xml emd-14522.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14522_fsc.xml emd_14522_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14522.png emd_14522.png | 56.8 KB | ||
| Masks |  emd_14522_msk_1.map emd_14522_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14522.cif.gz emd-14522.cif.gz | 6.4 KB | ||
| Others |  emd_14522_half_map_1.map.gz emd_14522_half_map_1.map.gz emd_14522_half_map_2.map.gz emd_14522_half_map_2.map.gz | 65.3 MB 65.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14522 http://ftp.pdbj.org/pub/emdb/structures/EMD-14522 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14522 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14522 | HTTPS FTP |
-Validation report
| Summary document |  emd_14522_validation.pdf.gz emd_14522_validation.pdf.gz | 822.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14522_full_validation.pdf.gz emd_14522_full_validation.pdf.gz | 821.9 KB | Display | |
| Data in XML |  emd_14522_validation.xml.gz emd_14522_validation.xml.gz | 16.9 KB | Display | |
| Data in CIF |  emd_14522_validation.cif.gz emd_14522_validation.cif.gz | 22.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14522 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14522 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14522 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14522 | HTTPS FTP |
-Related structure data
| Related structure data |  7z5jMC  7zqqC  7zr4C  7zseC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14522.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14522.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14522_msk_1.map emd_14522_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14522_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14522_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : MoSto dodecamer 2 x (AB)3
+Supramolecule #1: MoSto dodecamer 2 x (AB)3
+Macromolecule #1: Molybdenum storage protein subunit beta
+Macromolecule #2: Molybdenum storage protein subunit alpha
+Macromolecule #3: MOLYBDATE ION
+Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #5: MAGNESIUM ION
+Macromolecule #6: W11-O35 cluster
+Macromolecule #7: 1,1,3,3,5,7,7,9,11,15,15-undecakis($l^{1}-oxidanyl)-2$l^{4},4$l^{...
+Macromolecule #8: W8-O26 cluster
+Macromolecule #9: W10-O37 cluster
+Macromolecule #10: tungstate cluster
+Macromolecule #11: W3-O10 cluster
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 42.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)