[English] 日本語
 Yorodumi
Yorodumi- EMDB-14145: Structure of Native, iodinated bovine thyroglobulin solved on str... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Native, iodinated bovine thyroglobulin solved on strepavidin affinity grids. | |||||||||
 Map data Map data | Main map. This is a composit map build from a C2 map and two C1 map corresponding to both asymmetric units. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Thyroglobulin / thyroid hormone / iodine / precursor / HORMONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationhormone biosynthetic process / thyroid hormone generation / hormone activity / histone binding / extracellular space Similarity search - Function | |||||||||
| Biological species |  | |||||||||
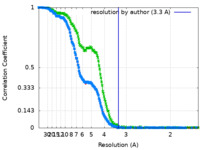
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Marechal N / Weitz JC / Serrano BP / Zhang X | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Formation of thyroid hormone revealed by a cryo-EM structure of native bovine thyroglobulin. Authors: Nils Marechal / Banyuhay P Serrano / Xinyan Zhang / Charles J Weitz /   Abstract: Thyroid hormones are essential regulators of metabolism, development, and growth. They are formed from pairs of iodinated tyrosine residues within the precursor thyroglobulin (TG), a 660-kDa ...Thyroid hormones are essential regulators of metabolism, development, and growth. They are formed from pairs of iodinated tyrosine residues within the precursor thyroglobulin (TG), a 660-kDa homodimer of the thyroid gland, by an oxidative coupling reaction. Tyrosine pairs that give rise to thyroid hormones have been assigned within the structure of human TG, but the process of hormone formation is poorly understood. Here we report a ~3.3-Å cryo-EM structure of native bovine TG with nascent thyroid hormone formed at one of the predicted hormonogenic sites. Local structural rearrangements provide insight into mechanisms underlying thyroid hormone formation and stabilization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14145.map.gz emd_14145.map.gz | 423.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14145-v30.xml emd-14145-v30.xml emd-14145.xml emd-14145.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14145_fsc.xml emd_14145_fsc.xml emd_14145_fsc_2.xml emd_14145_fsc_2.xml emd_14145_fsc_3.xml emd_14145_fsc_3.xml | 23.4 KB 23.3 KB 23.4 KB | Display Display Display |  FSC data file FSC data file |
| Images |  emd_14145.png emd_14145.png | 69.9 KB | ||
| Filedesc metadata |  emd-14145.cif.gz emd-14145.cif.gz | 7.7 KB | ||
| Others |  emd_14145_additional_1.map.gz emd_14145_additional_1.map.gz | 19.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14145 http://ftp.pdbj.org/pub/emdb/structures/EMD-14145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14145 | HTTPS FTP |
-Related structure data
| Related structure data |  7qtqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14145.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14145.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map. This is a composit map build from a C2 map and two C1 map corresponding to both asymmetric units. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Density modified map for the single asymmetric unit processing
| File | emd_14145_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified map for the single asymmetric unit processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Native bovine thyroglobulin from thyroid gland, containing T3/T4 ...
| Entire | Name: Native bovine thyroglobulin from thyroid gland, containing T3/T4 hormone precursors in two homonogenic sites. |
|---|---|
| Components |
|
-Supramolecule #1: Native bovine thyroglobulin from thyroid gland, containing T3/T4 ...
| Supramolecule | Name: Native bovine thyroglobulin from thyroid gland, containing T3/T4 hormone precursors in two homonogenic sites. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 670 KDa |
-Macromolecule #1: Thyroglobulin
| Macromolecule | Name: Thyroglobulin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 304.042562 KDa |
| Sequence | String: MALALWVFGL LDLICLASAN IFEYQVDAQP LRPCELQRER AFLKREDYVP QCAEDGSFQT VQCGKDGASC WCVDADGREV PGSRQPGRP AACLSFCQLQ KQQILLSSYI NSTATSYLPQ CQDSGDYSPV QCDLRRRQCW CVDAEGMEVY GTRQQGRPAR C PRSCEIRN ...String: MALALWVFGL LDLICLASAN IFEYQVDAQP LRPCELQRER AFLKREDYVP QCAEDGSFQT VQCGKDGASC WCVDADGREV PGSRQPGRP AACLSFCQLQ KQQILLSSYI NSTATSYLPQ CQDSGDYSPV QCDLRRRQCW CVDAEGMEVY GTRQQGRPAR C PRSCEIRN RRLLHGVGDR SPPQCSPDGA FRPVQCKFVN TTDMMIFDLV HSYSRFPDAF VTFSSFRSRF PEVSGYCYCA DS QGRELAE TGLELLLDEI YDTIFAGLDL ASTFAETTLY RILQRRFLAV QLVISGRFRC PTKCEVERFA ATSFRHPYVP SCH PDGEYQ AAQCQQGGPC WCVDSRGQEI PGTRQRGEPP SCAEDQSCPS ERRRAFSRLR FGPSGYFSRR SLLLAPEEGP VSQR FARFT ASCPPSIKEL FLDSGIFQPM LQGRDTRFVA PESLKEAIRG LFPSRELARL ALQFTTNAKR LQQNLFGGRF LVKVG QFNL SGALGTRGTF NFSHFFQQLG LPGFQDGRAL ADLAKPLSVG LNSNPASEAP KASKIDVALR KPVVGSFGFE VNLQEN QNA LQFLSSFLEL PEFLLFLQHA ISVPEDIARD LGDVMEMVFS SQGCGQAPGS LFVPACTAEG SYEEVQCFAG DCWCVDA QG RELAGSRVRG GRPRCPTECE KQRARMQSLL GSQPAGSSLF VPACTSKGNF LPVQCFNSEC YCVDTEGQPI PGTRSALG E PKKCPSPCQL QAERAFLGTV RTLVSNPSTL PALSSIYIPQ CSASGQWSPV QCDGPPEQAF EWYERWEAQN SAGQALTPA ELLMKIMSYR EAASRNFRLF IQNLYEAGQQ GIFPGLARYS SFQDVPVSVL EGNQTQPGGN VFLEPYLFWQ ILNGQLDRYP GPYSDFSAP LAHFDLRSCW CVDEAGQKLE GTRNEPNKVP ACPGSCEEVK LRVLQFIREA EEIVTYSNSS RFPLGESFLA A KGIRLTDE ELAFPPLSPS RETFLEKFLS GSDYAIRLAA QSTFDFYQRR LVTLAESPRA PSPVWSSAYL PQCDAFGGWE PV QCHAATG HCWCVDGKGE YVPTSLTARS RQIPQCPTSC ERLRASGLLS SWKQAGVQAE PSPKDLFIPT CLETGEFARL QAS EAGTWC VDPASGEGVP PGTNSSAQCP SLCEVLQSGV PSRRTSPGYS PACRAEDGGF SPVQCDPAQG SCWCVLGSGE EVPG TRVAG SQPACESPQC PLPFSVADVA GGAILCERAS GLGAAAGQRC QLRCSQGYRS AFPPEPLLCS VQRRRWESRP PQPRA CQRP QFWQTLQTQA QFQLLLPLGK VCSADYSGLL LAFQVFLLDE LTARGFCQIQ VKTAGTPVSI PVCDDSSVKV ECLSRE RLG VNITWKLQLV DAPPASLPDL QDVEEALAGK YLAGRFADLI QSGTFQLHLD SKTFSADTSI RFLQGDRFGT SPRTQFG CL EGFGRVVAAS DASQDALGCV KCPEGSYFQD EQCIPCPAGF YQEQAGSLAC VPCPEGRTTV YAGAFSQTHC VTDCQKNE V GLQCDQDSQY RASQRDRTSG KAFCVDGEGR RLPWTEAEAP LVDAQCLVMR KFEKLPESKV IFSADVAVMV RSEVPGSES SLMQCLADCA LDEACGFLTV STAGSEVSCD FYAWASDSIA CTTSGRSEDA LGTSQATSFG SLQCQVKVRS REGDPLAVYL KKGQEFTIT GQKRFEQTGF QSALSGMYSP VTFSASGASL AEVHLFCLLA CDHDSCCDGF ILVQVQGGPL LCGLLSSPDV L LCHVRDWR DPAEAQANAS CPGVTYDQDS RQVTLRLGGQ EIRGLTPLEG TQDTLTSFQQ VYLWKDSDMG SRSESMGCRR DT EPRPASP SETDLTTGLF SPVDLIQVIV DGNVSLPSQQ HWLFKHLFSL QQANLWCLSR CAGEPSFCQL AEVTDSEPLY FTC TLYPEA QVCDDILESS PKGCRLILPR RPSALYRKKV VLQDRVKNFY NRLPFQKLTG ISIRNKVPMS DKSISSGFFE CERL CDMDP CCTGFGFLNV SQLKGGEVTC LTLNSLGLQT CSEEYGGVWR ILDCGSPDTE VRTYPFGWYQ KPVSPSDAPS FCPSV ALPA LTENVALDSW QSLALSSVIV DPSIRNFDVA HISTAAVGNF SAARDRCLWE CSRHQDCLVT TLQTQPGAVR CMFYAD TQS CTHSLQAQNC RLLLHEEATY IYRKPNIPLP GFGTSSPSVP IATHGQLLGR SQAIQVGTSW KPVDQFLGVP YAAPPLG EK RFRAPEHLNW TGSWEATKPR ARCWQPGIRT PTPPGVSEDC LYLNVFVPQN MAPNASVLVF FHNAAEGKGS GDRPAVDG S FLAAVGNLIV VTASYRTGIF GFLSSGSSEL SGNWGLLDQV VALTWVQTHI QAFGGDPRRV TLAADRGGAD IASIHLVTT RAANSRLFRR AVLMGGSALS PAAVIRPERA RQQAAALAKE VGCPSSSVQE MVSCLRQEPA RILNDAQTKL LAVSGPFHYW GPVVDGQYL RETPARVLQR APRVKVDLLI GSSQDDGLIN RAKAVKQFEE SQGRTSSKTA F(DHA)QALQNSLG GEAADAG VQ AAATWYYSLE HDSDD(T44)ASFS RALEQATRDY FIICPVIDMA SHWARTVRGN VFMYHAPESY SHSSLELLTD VLYA FGLPF YPAYEGQFTL EEKSLSLKIM QYFSNFIRSG NPNYPHEFSR RAPEFAAPWP DFVPRDGAES YKELSVLLPN RQGLK KADC SFWSKYIQSL KASADETKDG PSADSEEEDQ PAGSGLTEDL LGLPELASKT YSK UniProtKB: Thyroglobulin |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 4 Details: Streaptavidin 2D crystal supported by a biotinylated monolayer, stabilized by thin carbon evaporation on lipid tails | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Number real images: 34597 / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7qtq: |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)