[English] 日本語
 Yorodumi
Yorodumi- EMDB-14002: Specific features and methylation sites of a plant ribosome. 40S ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Specific features and methylation sites of a plant ribosome. 40S body ribosomal subunit. | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Solanum lycopersicum / cytosolic ribosome / 80S / plant / rRNA / RIBOSOME | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationendonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / chloroplast / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / small-subunit processome / rRNA processing / ribosomal small subunit biogenesis / small ribosomal subunit rRNA binding / ribosomal small subunit assembly / cytosolic small ribosomal subunit ...endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / chloroplast / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / small-subunit processome / rRNA processing / ribosomal small subunit biogenesis / small ribosomal subunit rRNA binding / ribosomal small subunit assembly / cytosolic small ribosomal subunit / cytoplasmic translation / cytosolic large ribosomal subunit / rRNA binding / ribosome / structural constituent of ribosome / translation / ribonucleoprotein complex / mRNA binding / nucleolus / RNA binding / metal ion binding / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
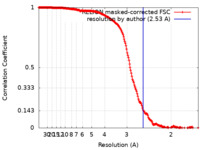
| Method | single particle reconstruction / cryo EM / Resolution: 2.53 Å | |||||||||||||||
 Authors Authors | Cottilli P / Itoh Y / Amunts A | |||||||||||||||
| Funding support | European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Plant Commun / Year: 2022 Journal: Plant Commun / Year: 2022Title: Cryo-EM structure and rRNA modification sites of a plant ribosome. Authors: Patrick Cottilli / Yuzuru Itoh / Yuko Nobe / Anton S Petrov / Purificación Lisón / Masato Taoka / Alexey Amunts /     Abstract: Protein synthesis in crop plants contributes to the balance of food and fuel on our planet, which influences human metabolic activity and lifespan. Protein synthesis can be regulated with respect to ...Protein synthesis in crop plants contributes to the balance of food and fuel on our planet, which influences human metabolic activity and lifespan. Protein synthesis can be regulated with respect to changing environmental cues via the deposition of chemical modifications into rRNA. Here, we present the structure of a plant ribosome from tomato and a quantitative mass spectrometry analysis of its rRNAs. The study reveals fine features of the ribosomal proteins and 71 plant-specific rRNA modifications, and it re-annotates 30 rRNA residues in the available sequence. At the protein level, isoAsp is found in position 137 of uS11, and a zinc finger previously believed to be universal is missing from eL34, suggesting a lower effect of zinc deficiency on protein synthesis in plants. At the rRNA level, the plant ribosome differs markedly from its human counterpart with respect to the spatial distribution of modifications. Thus, it represents an additional layer of gene expression regulation, highlighting the molecular signature of a plant ribosome. The results provide a reference model of a plant ribosome for structural studies and an accurate marker for molecular ecology. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14002.map.gz emd_14002.map.gz | 330 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14002-v30.xml emd-14002-v30.xml emd-14002.xml emd-14002.xml | 38.9 KB 38.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14002_fsc.xml emd_14002_fsc.xml | 19.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14002.png emd_14002.png | 26.4 KB | ||
| Masks |  emd_14002_msk_1.map emd_14002_msk_1.map | 600.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14002.cif.gz emd-14002.cif.gz | 9.9 KB | ||
| Others |  emd_14002_half_map_1.map.gz emd_14002_half_map_1.map.gz emd_14002_half_map_2.map.gz emd_14002_half_map_2.map.gz | 482.7 MB 482.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14002 http://ftp.pdbj.org/pub/emdb/structures/EMD-14002 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14002 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14002 | HTTPS FTP |
-Validation report
| Summary document |  emd_14002_validation.pdf.gz emd_14002_validation.pdf.gz | 853.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14002_full_validation.pdf.gz emd_14002_full_validation.pdf.gz | 853.1 KB | Display | |
| Data in XML |  emd_14002_validation.xml.gz emd_14002_validation.xml.gz | 27 KB | Display | |
| Data in CIF |  emd_14002_validation.cif.gz emd_14002_validation.cif.gz | 36 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14002 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14002 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14002 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14002 | HTTPS FTP |
-Related structure data
| Related structure data |  7qixMC  7qiwC  7qiyC  7qizC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14002.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14002.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
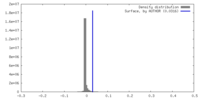
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14002_msk_1.map emd_14002_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
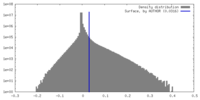
| Density Histograms |
-Half map: #1
| File | emd_14002_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
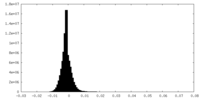
| Density Histograms |
-Half map: #2
| File | emd_14002_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 40S body ribosomal subunit
+Supramolecule #1: 40S body ribosomal subunit
+Macromolecule #1: 18S rRNA body
+Macromolecule #2: 60S ribosomal protein L41
+Macromolecule #3: 40S ribosomal protein SA
+Macromolecule #4: 40S ribosomal protein S3a
+Macromolecule #5: 40S body ribosomal protein eS4
+Macromolecule #6: 40S ribosomal protein S7
+Macromolecule #7: 40S ribosomal protein S8
+Macromolecule #8: Ribosomal_S17_N domain-containing protein
+Macromolecule #9: 40S ribosomal protein S17
+Macromolecule #10: 40S ribosomal protein S21
+Macromolecule #11: 40S body ribosomal protein uS12
+Macromolecule #12: 40S ribosomal protein S26
+Macromolecule #13: S5 DRBM domain-containing protein
+Macromolecule #14: 40S ribosomal protein S6
+Macromolecule #15: 40S body ribosomal protein uS4
+Macromolecule #16: 30S ribosomal protein S15, chloroplastic
+Macromolecule #17: Ribosomal protein S14
+Macromolecule #18: 40S ribosomal protein S24
+Macromolecule #19: 40S ribosomal protein S27
+Macromolecule #20: 40S ribosomal protein S30
+Macromolecule #21: 40S ribosomal protein S15a-1
+Macromolecule #22: Ribosomal protein L19
+Macromolecule #23: POTASSIUM ION
+Macromolecule #24: MAGNESIUM ION
+Macromolecule #25: ZINC ION
+Macromolecule #26: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


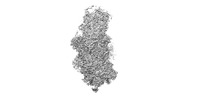










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)