+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Leishmania major actin filament in ADP-Pi state | |||||||||
 Map data Map data | Real-space symmetrized and Phenix autosharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Actin / Filament / Parasite / ADP-Pi / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationkinetoplast / actin cytoskeleton / endonuclease activity / chromatin remodeling / ATP binding Similarity search - Function | |||||||||
| Biological species |  Leishmania major (eukaryote) Leishmania major (eukaryote) | |||||||||
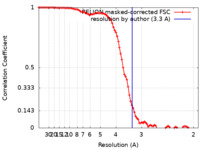
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Kotila T / Muniyandi S / Lappalainen P / Huiskonen JT | |||||||||
| Funding support |  Finland, 2 items Finland, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite. Authors: Tommi Kotila / Hugo Wioland / Muniyandi Selvaraj / Konstantin Kogan / Lina Antenucci / Antoine Jégou / Juha T Huiskonen / Guillaume Romet-Lemonne / Pekka Lappalainen /   Abstract: Actin polymerization generates forces for cellular processes throughout the eukaryotic kingdom, but our understanding of the 'ancient' actin turnover machineries is limited. We show that, ...Actin polymerization generates forces for cellular processes throughout the eukaryotic kingdom, but our understanding of the 'ancient' actin turnover machineries is limited. We show that, despite > 1 billion years of evolution, pathogenic Leishmania major parasite and mammalian actins share the same overall fold and co-polymerize with each other. Interestingly, Leishmania harbors a simple actin-regulatory machinery that lacks cofilin 'cofactors', which accelerate filament disassembly in higher eukaryotes. By applying single-filament biochemistry we discovered that, compared to mammalian proteins, Leishmania actin filaments depolymerize more rapidly from both ends, and are severed > 100-fold more efficiently by cofilin. Our high-resolution cryo-EM structures of Leishmania ADP-, ADP-Pi- and cofilin-actin filaments identify specific features at actin subunit interfaces and cofilin-actin interactions that explain the unusually rapid dynamics of parasite actin filaments. Our findings reveal how divergent parasites achieve rapid actin dynamics using a remarkably simple set of actin-binding proteins, and elucidate evolution of the actin cytoskeleton. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13863.map.gz emd_13863.map.gz | 54.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13863-v30.xml emd-13863-v30.xml emd-13863.xml emd-13863.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13863_fsc.xml emd_13863_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13863.png emd_13863.png | 98.9 KB | ||
| Masks |  emd_13863_msk_1.map emd_13863_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13863.cif.gz emd-13863.cif.gz | 6.9 KB | ||
| Others |  emd_13863_additional_1.map.gz emd_13863_additional_1.map.gz emd_13863_half_map_1.map.gz emd_13863_half_map_1.map.gz emd_13863_half_map_2.map.gz emd_13863_half_map_2.map.gz | 59.7 MB 49.4 MB 49.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13863 http://ftp.pdbj.org/pub/emdb/structures/EMD-13863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13863 | HTTPS FTP |
-Related structure data
| Related structure data |  7q8bMC  7q8cC  7q8sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13863.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13863.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Real-space symmetrized and Phenix autosharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
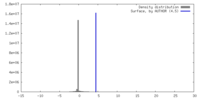
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13863_msk_1.map emd_13863_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
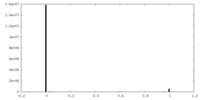
| Density Histograms |
-Additional map: Postprocess from Relion
| File | emd_13863_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess from Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
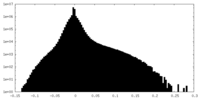
| Density Histograms |
-Half map: Half-map 2
| File | emd_13863_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
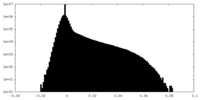
| Density Histograms |
-Half map: Half-map 1
| File | emd_13863_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Polymerized actin
| Entire | Name: Polymerized actin |
|---|---|
| Components |
|
-Supramolecule #1: Polymerized actin
| Supramolecule | Name: Polymerized actin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Leishmania major (eukaryote) Leishmania major (eukaryote) |
-Macromolecule #1: Actin
| Macromolecule | Name: Actin / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leishmania major (eukaryote) Leishmania major (eukaryote) |
| Molecular weight | Theoretical: 42.063867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADNEQSSIV CDNGSGMVKA GFSGDDAPRH VFPSIVGRPK NMQAMMGSAN KTVYVGDEAQ SKRGVLSLKY PIEHGIVTNW DDMEKIWHH TFYNELRVNP EQHNVLLTEA PMNPKQNREK MTQIMFETFN VPSLYIGIQA VLSLYSSGRT TGIVLDAGDG V THTVPIYE ...String: MADNEQSSIV CDNGSGMVKA GFSGDDAPRH VFPSIVGRPK NMQAMMGSAN KTVYVGDEAQ SKRGVLSLKY PIEHGIVTNW DDMEKIWHH TFYNELRVNP EQHNVLLTEA PMNPKQNREK MTQIMFETFN VPSLYIGIQA VLSLYSSGRT TGIVLDAGDG V THTVPIYE GYSLPHAVRR VDMAGRDLTE YLMKIMMETG TTFTTTAEKE IVRNVKEQLC YVALDFEEEM TNSAKSANEE AF ELPDGNV MMVGNQRFRC PEVLFKPSLI GLDEAPGFPE MVYQSINKCD IDVRRELYGN IVLSGGSTMF LNLPERLAKE ISN LAPSSI KPKVVAPPER KYSVWIGGSI LSSLTTFQTM WVKKSEYDES GPSIVHNKCF UniProtKB: Actin |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 4 / Number of copies: 5 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.2 Details: Phosphate buffered saline supplemented with 0.2 mM ATP, 1 mM MgCl2, 0.4 mM EGTA and 1 mM DTT. |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 279.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 45 / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-7q8b: |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)