[English] 日本語
 Yorodumi
Yorodumi- EMDB-13609: Structure of CtAtm1 in the inward-open with Glutathione-complexed... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of CtAtm1 in the inward-open with Glutathione-complexed [2Fe-2S] cluster bound | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ABC exporter / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type transporter activity / mitochondrial inner membrane / ATP hydrolysis activity / ATP binding Similarity search - Function | ||||||||||||
| Biological species |  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) | ||||||||||||
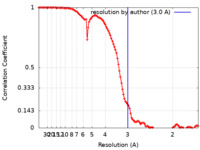
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Li P / Wang KT | ||||||||||||
| Funding support |  Denmark, 3 items Denmark, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structures of Atm1 provide insight into [2Fe-2S] cluster export from mitochondria. Authors: Ping Li / Amber L Hendricks / Yong Wang / Rhiza Lyne E Villones / Karin Lindkvist-Petersson / Gabriele Meloni / J A Cowan / Kaituo Wang / Pontus Gourdon /     Abstract: In eukaryotes, iron-sulfur clusters are essential cofactors for numerous physiological processes, but these clusters are primarily biosynthesized in mitochondria. Previous studies suggest ...In eukaryotes, iron-sulfur clusters are essential cofactors for numerous physiological processes, but these clusters are primarily biosynthesized in mitochondria. Previous studies suggest mitochondrial ABCB7-type exporters are involved in maturation of cytosolic iron-sulfur proteins. However, the molecular mechanism for how the ABCB7-type exporters participate in this process remains elusive. Here, we report a series of cryo-electron microscopy structures of a eukaryotic homolog of human ABCB7, CtAtm1, determined at average resolutions ranging from 2.8 to 3.2 Å, complemented by functional characterization and molecular docking in silico. We propose that CtAtm1 accepts delivery from glutathione-complexed iron-sulfur clusters. A partially occluded state links cargo-binding to residues at the mitochondrial matrix interface that line a positively charged cavity, while the binding region becomes internalized and is partially divided in an early occluded state. Collectively, our findings substantially increase the understanding of the transport mechanism of eukaryotic ABCB7-type proteins. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13609.map.gz emd_13609.map.gz | 64.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13609-v30.xml emd-13609-v30.xml emd-13609.xml emd-13609.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13609_fsc.xml emd_13609_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_13609.png emd_13609.png | 42.5 KB | ||
| Filedesc metadata |  emd-13609.cif.gz emd-13609.cif.gz | 5.7 KB | ||
| Others |  emd_13609_half_map_1.map.gz emd_13609_half_map_1.map.gz emd_13609_half_map_2.map.gz emd_13609_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13609 http://ftp.pdbj.org/pub/emdb/structures/EMD-13609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13609 | HTTPS FTP |
-Validation report
| Summary document |  emd_13609_validation.pdf.gz emd_13609_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13609_full_validation.pdf.gz emd_13609_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_13609_validation.xml.gz emd_13609_validation.xml.gz | 19.3 KB | Display | |
| Data in CIF |  emd_13609_validation.cif.gz emd_13609_validation.cif.gz | 24.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13609 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13609 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13609 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13609 | HTTPS FTP |
-Related structure data
| Related structure data |  7proMC  7pqxC  7pr1C  7pruC  7psdC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13609.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13609.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
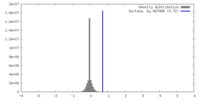
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_13609_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
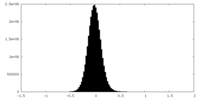
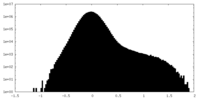
| Density Histograms |
-Half map: #2
| File | emd_13609_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
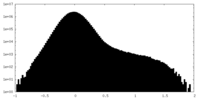
| Density Histograms |
- Sample components
Sample components
-Entire : atm1
| Entire | Name: atm1 |
|---|---|
| Components |
|
-Supramolecule #1: atm1
| Supramolecule | Name: atm1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
-Macromolecule #1: Putative iron-sulfur protein
| Macromolecule | Name: Putative iron-sulfur protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 77.199812 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSPRSRLLAS SLRGGLLLRP CSARRSTLLN PSPSPLRKAI FHTTSLRAFK NLAEIATSSK EDASSLKPKG KPSPPGDPLA TIEKTAAEQ RRADWAIIKQ MSRYLWPKDS WSDKARVLLA LSLLVGGKVL NVHVPFYFRE IIDRLNIDVA AVGGTVSAVA G AVIFAYGA ...String: MSPRSRLLAS SLRGGLLLRP CSARRSTLLN PSPSPLRKAI FHTTSLRAFK NLAEIATSSK EDASSLKPKG KPSPPGDPLA TIEKTAAEQ RRADWAIIKQ MSRYLWPKDS WSDKARVLLA LSLLVGGKVL NVHVPFYFRE IIDRLNIDVA AVGGTVSAVA G AVIFAYGA SRIGAVVSQE LRNAVFSSVA QKAIRRVATQ TFGHLLNLDL SFHLSKQTGG LTRAIDRGTK GISYLLTSMV FH IVPTALE IGMVCGILTY QFGWEFAAIT AATMAAYTAF TITTTAWRTK FRRQANAADN AASTVAVDSL INYEAVKYFN NEA YEIARY DKALQAYERS SIKVATSLAF LNSGQNIIFS SALTLMMWLG ARGVLAGDLS VGDLVLINQL VFQLSVPLNF LGSV YRELR QSLLDMETLF DLQKVNVTIR EAPNAKPLAL PKGGEIRFEN VTFGYYPDRP ILRNLSLTIP AGKKVAVVGP SGCGK STLL RLLFRSYDPQ QGKIFIDDQD IKSVTLESLR KSIGVVPQDT PLFNDTVELN IRYGNVNATQ EQVIAAAQKA HIHEKI ISW PHGYQTRVGE RGLMISGGEK QRLAVSRLIL KDPPLLFFDQ ATSALDTHTE QALMANINEV VKEKKRTALF VAHRLRT IY DADLIIVLKE GVVVEQGSHR ELMERDGVYA ELWMAQEMLH QGEAAEEPAE GEKAEEKK UniProtKB: Iron-sulfur clusters transporter ATM1, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)