[English] 日本語
 Yorodumi
Yorodumi- EMDB-13563: Human coronavirus OC43 spike glycoprotein ectodomain in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 47C9 antibody Fab fragment | |||||||||
 Map data Map data | Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 47C9 antibody Fab fragment | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Coronavirus / Glycoprotein / Antibody / Spike / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Human coronavirus OC43 / Human coronavirus OC43 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
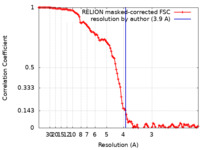
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Hesketh EL / Townend S / Ranson NA / Hurdiss DL | |||||||||
| Funding support |  Netherlands, Netherlands,  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Antigenic structure of the human coronavirus OC43 spike reveals exposed and occluded neutralizing epitopes. Authors: Chunyan Wang / Emma L Hesketh / Tatiana M Shamorkina / Wentao Li / Peter J Franken / Dubravka Drabek / Rien van Haperen / Sarah Townend / Frank J M van Kuppeveld / Frank Grosveld / Neil A ...Authors: Chunyan Wang / Emma L Hesketh / Tatiana M Shamorkina / Wentao Li / Peter J Franken / Dubravka Drabek / Rien van Haperen / Sarah Townend / Frank J M van Kuppeveld / Frank Grosveld / Neil A Ranson / Joost Snijder / Raoul J de Groot / Daniel L Hurdiss / Berend-Jan Bosch /    Abstract: Human coronavirus OC43 is a globally circulating common cold virus sustained by recurrent reinfections. How it persists in the population and defies existing herd immunity is unknown. Here we focus ...Human coronavirus OC43 is a globally circulating common cold virus sustained by recurrent reinfections. How it persists in the population and defies existing herd immunity is unknown. Here we focus on viral glycoprotein S, the target for neutralizing antibodies, and provide an in-depth analysis of its antigenic structure. Neutralizing antibodies are directed to the sialoglycan-receptor binding site in S1 domain, but, remarkably, also to S1. The latter block infection yet do not prevent sialoglycan binding. While two distinct neutralizing S1 epitopes are readily accessible in the prefusion S trimer, other sites are occluded such that their accessibility must be subject to conformational changes in S during cell-entry. While non-neutralizing antibodies were broadly reactive against a collection of natural OC43 variants, neutralizing antibodies generally displayed restricted binding breadth. Our data provide a structure-based understanding of protective immunity and adaptive evolution for this endemic coronavirus which emerged in humans long before SARS-CoV-2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13563.map.gz emd_13563.map.gz | 111.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13563-v30.xml emd-13563-v30.xml emd-13563.xml emd-13563.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13563_fsc.xml emd_13563_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_13563.png emd_13563.png | 73.3 KB | ||
| Filedesc metadata |  emd-13563.cif.gz emd-13563.cif.gz | 7.6 KB | ||
| Others |  emd_13563_additional_1.map.gz emd_13563_additional_1.map.gz emd_13563_half_map_1.map.gz emd_13563_half_map_1.map.gz emd_13563_half_map_2.map.gz emd_13563_half_map_2.map.gz | 98.7 MB 99.2 MB 99.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13563 http://ftp.pdbj.org/pub/emdb/structures/EMD-13563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13563 | HTTPS FTP |
-Related structure data
| Related structure data |  7po5MC  7pnmC  7pnqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13563.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13563.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 47C9 antibody Fab fragment | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_13563_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_13563_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_13563_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human coronavirus OC43 spike glycoprotein ectodomain in complex w...
| Entire | Name: Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 47C9 antibody Fab fragment |
|---|---|
| Components |
|
-Supramolecule #1: Human coronavirus OC43 spike glycoprotein ectodomain in complex w...
| Supramolecule | Name: Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 47C9 antibody Fab fragment type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Human coronavirus OC43 / Strain: USA/1967 Human coronavirus OC43 / Strain: USA/1967 |
| Molecular weight | Theoretical: 583 KDa |
-Macromolecule #1: 47C9 antibody heavy chain
| Macromolecule | Name: 47C9 antibody heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.521896 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQQWGAG LLKPSETLSL TCAVYGGSFS GYYWNWIRQP PGKGLEWIGE INHSGSTNYN PSLKSRVTIS VDTSKNQFSL KLSSVTAAD TAVYYCARGN YYGSGSYVDY GMDVWGQGTT VTVSS |
-Macromolecule #2: 47C9 antibody light chain
| Macromolecule | Name: 47C9 antibody light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.448732 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVMTQSPAT LSVSPGERAT LSCRASQSIS SNLAWYQQKP GQAPRLLIYG ASTRATGIPA RFSGSGSGTE FTLTISSLQS EDFAVYYCQ QSNNWPLTFG GGTKVEIK |
-Macromolecule #3: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human coronavirus OC43 Human coronavirus OC43 |
| Molecular weight | Theoretical: 146.738734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPMGSLQPLA TLYLLGMLVA SVLAVIGDLK CTSDNINDKD TGPPPISTDT VDVTNGLGTY YVLDRVYLNT TLFLNGYYPT SGSTYRNMA LKGSVLLSRL WFKPPFLSDF INGIFAKVKN TKVIKDRVMY SEFPAITIGS TFVNTSYSVV VQPRTINSTQ D GDNKLQGL ...String: MPMGSLQPLA TLYLLGMLVA SVLAVIGDLK CTSDNINDKD TGPPPISTDT VDVTNGLGTY YVLDRVYLNT TLFLNGYYPT SGSTYRNMA LKGSVLLSRL WFKPPFLSDF INGIFAKVKN TKVIKDRVMY SEFPAITIGS TFVNTSYSVV VQPRTINSTQ D GDNKLQGL LEVSVCQYNM CEYPQTICHP NLGNHRKELW HLDTGVVSCL YKRNFTYDVN ADYLYFHFYQ EGGTFYAYFT DT GVVTKFL FNVYLGMALS HYYVMPLTCN SKLTLEYWVT PLTSRQYLLA FNQDGIIFNA VDCMSDFMSE IKCKTQSIAP PTG VYELNG YTVQPIADVY RRKPNLPNCN IEAWLNDKSV PSPLNWERKT FSNCNFNMSS LMSFIQADSF TCNNIDAAKI YGMC FSSIT IDKFAIPNGR KVDLQLGNLG YLQSFNYRID TTATSCQLYY NLPAANVSVS RFNPSTWNKR FGFIEDSVFK PRPAG VLTN HDVVYAQHCF KAPKNFCPCK LNGSCVGSGP GKNNGIGTCP AGTNYLTCDN LCTPDPITFT GTYKCPQTKS LVGIGE HCS GLAVKSDYCG GNSCTCRPQA FLGWSADSCL QGDKCNIFAN FILHDVNSGL TCSTDLQKAN TDIILGVCVN YDLYGIL GQ GIFVEVNATY YNSWQNLLYD SNGNLYGFRD YITNRTFMIR SCYSGRVSAA FHANSSEPAL LFRNIKCNYV FNNSLTRQ L QPINYFDSYL GCVVNAYNST AISVQTCDLT VGSGYCVDYS KNRRSRGAIT TGYRFTNFEP FTVNSVNDSL EPVGGLYEI QIPSEFTIGN MVEFIQTSSP KVTIDCAAFV CGDYAACKSQ LVEYGSFCDN INAILTEVNE LLDTTQLQVA NSLMNGVTLS TKLKDGVNF NVDDINFSPV LGCLGSECSK ASSRSAIEDL LFDKVKLSDV GFVEAYNNCT GGAEIRDLIC VQSYKGIKVL P PLLSENQF SGYTLAATSA SLFPPWTAAA GVPFYLNVQY RINGLGVTMD VLSQNQKLIA NAFNNALYAI QEGFDATNSA LV KIQAVVN ANAEALNNLL QQLSNRFGAI SASLQEILSR LDALEAEAQI DRLINGRLTA LNAYVSQQLS DSTLVKFSAA QAM EKVNEC VKSQSSRINF CGNGNHIISL VQNAPYGLYF IHFSYVPTKY VTARVSPGLC IAGDRGIAPK SGYFVNVNNT WMYT GSGYY YPEPITENNV VVMSTCAVNY TKAPYVMLNT SIPNLPDFKE ELDQWFKNQT SVAPDLSLDY INVTFLDLLI KRMKQ IEDK IEEIESKQKK IENEIARIKK IKLVPRGSLE WSHPQFEK UniProtKB: Spike glycoprotein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Purified OC43 spike ectodomain and the antibody Fab fragments were incubated together for 5 minutes at a 1:1 molar ratio |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Details | A 30 degree stage tilt was employed during data collection to increase the number of side views visualised due to preferential orientation. |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 380 / Average exposure time: 60.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)