[English] 日本語
 Yorodumi
Yorodumi- EMDB-13257: TmHydABC- T. maritima bifurcating hydrogenase with bridge domain ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain closed | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Hydrogenase / bifurcation / confurcaction / cryoEM / electron transfer / OXIDOREDUCTASE / complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationhydrogenase (NAD+, ferredoxin) / NADH dehydrogenase (ubiquinone) activity / ATP synthesis coupled electron transport / 2 iron, 2 sulfur cluster binding / FMN binding / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / metal ion binding / membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   Thermotoga maritima MSB8 (bacteria) / Thermotoga maritima MSB8 (bacteria) /   Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) (bacteria) Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) (bacteria) | ||||||||||||
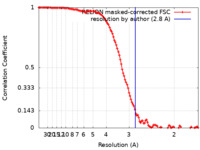
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | ||||||||||||
 Authors Authors | Furlan C / Chongdar N | ||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Germany, Germany,  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase. Authors: Chris Furlan / Nipa Chongdar / Pooja Gupta / Wolfgang Lubitz / Hideaki Ogata / James N Blaza / James A Birrell /    Abstract: Electron bifurcation is a fundamental energy conservation mechanism in nature in which two electrons from an intermediate-potential electron donor are split so that one is sent along a high-potential ...Electron bifurcation is a fundamental energy conservation mechanism in nature in which two electrons from an intermediate-potential electron donor are split so that one is sent along a high-potential pathway to a high-potential acceptor and the other is sent along a low-potential pathway to a low-potential acceptor. This process allows endergonic reactions to be driven by exergonic ones and is an alternative, less recognized, mechanism of energy coupling to the well-known chemiosmotic principle. The electron-bifurcating [FeFe] hydrogenase from (HydABC) requires both NADH and ferredoxin to reduce protons generating hydrogen. The mechanism of electron bifurcation in HydABC remains enigmatic in spite of intense research efforts over the last few years. Structural information may provide the basis for a better understanding of spectroscopic and functional information. Here, we present a 2.3 Å electron cryo-microscopy structure of HydABC. The structure shows a heterododecamer composed of two independent 'halves' each made of two strongly interacting HydABC heterotrimers connected via a [4Fe-4S] cluster. A central electron transfer pathway connects the active sites for NADH oxidation and for proton reduction. We identified two conformations of a flexible iron-sulfur cluster domain: a 'closed bridge' and an 'open bridge' conformation, where a Zn site may act as a 'hinge' allowing domain movement. Based on these structural revelations, we propose a possible mechanism of electron bifurcation in HydABC where the flavin mononucleotide serves a dual role as both the electron bifurcation center and as the NAD reduction/NADH oxidation site. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13257.map.gz emd_13257.map.gz | 12.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13257-v30.xml emd-13257-v30.xml emd-13257.xml emd-13257.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13257_fsc.xml emd_13257_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13257.png emd_13257.png | 130.6 KB | ||
| Masks |  emd_13257_msk_1.map emd_13257_msk_1.map | 134.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13257.cif.gz emd-13257.cif.gz | 7.2 KB | ||
| Others |  emd_13257_additional_1.map.gz emd_13257_additional_1.map.gz emd_13257_half_map_1.map.gz emd_13257_half_map_1.map.gz emd_13257_half_map_2.map.gz emd_13257_half_map_2.map.gz | 11.8 MB 106.2 MB 106.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13257 http://ftp.pdbj.org/pub/emdb/structures/EMD-13257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13257 | HTTPS FTP |
-Related structure data
| Related structure data |  7p91MC  7p5hC  7p8nC  7p92C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13257.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13257.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13257_msk_1.map emd_13257_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_13257_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13257_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13257_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer of TmHydABC protomers in a bridge closed conformation
| Entire | Name: Dimer of TmHydABC protomers in a bridge closed conformation |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of TmHydABC protomers in a bridge closed conformation
| Supramolecule | Name: Dimer of TmHydABC protomers in a bridge closed conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima MSB8 (bacteria) Thermotoga maritima MSB8 (bacteria) |
| Molecular weight | Theoretical: 640 KDa |
-Macromolecule #1: Fe-hydrogenase, subunit alpha
| Macromolecule | Name: Fe-hydrogenase, subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: hydrogenase (NAD+, ferredoxin) |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) (bacteria) Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) (bacteria)Strain: ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8 |
| Molecular weight | Theoretical: 72.351391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKIYVDGREV IINDNERNLL EALKNVGIEI PNLCYLSEAS IYGACRMCLV EINGQITTSC TLKPYEGMKV KTNTPEIYEM RRNILELIL ATHNRDCTTC DRNGSCKLQK YAEDFGIRKI RFEALKKEHV RDESAPVVRD TSKCILCGDC VRVCEEIQGV G VIEFAKRG ...String: MKIYVDGREV IINDNERNLL EALKNVGIEI PNLCYLSEAS IYGACRMCLV EINGQITTSC TLKPYEGMKV KTNTPEIYEM RRNILELIL ATHNRDCTTC DRNGSCKLQK YAEDFGIRKI RFEALKKEHV RDESAPVVRD TSKCILCGDC VRVCEEIQGV G VIEFAKRG FESVVTTAFD TPLIETECVL CGQCVAYCPT GALSIRNDID KLIEALESDK IVIGMIAPAV RAAIQEEFGI DE DVAMAEK LVSFLKTIGF DKVFDVSFGA DLVAYEEAHE FYERLKKGER LPQFTSCCPA WVKHAEHTYP QYLQNLSSVK SPQ QALGTV IKKIYARKLG VPEEKIFLVS FMPCTAKKFE AEREEHEGIV DIVLTTRELA QLIKMSRIDI NRVEPQPFDR PYGV SSQAG LGFGKAGGVF SCVLSVLNEE IGIEKVDVKS PEDGIRVAEV TLKDGTSFKG AVIYGLGKVK KFLEERKDVE IIEVM ACNY GCVGGGGQPY PNDSRIREHR AKVLRDTMGI KSLLTPVENL FLMKLYEEDL KDEHTRHEIL HTTYRPRRRY PEKDVE ILP VPNGEKRTVK VCLGTSCYTK GSYEILKKLV DYVKENDMEG KIEVLGTFCV ENCGASPNVI VDDKIIGGAT FEKVLEE LS KNG UniProtKB: Bifurcating [FeFe] hydrogenase alpha subunit |
-Macromolecule #2: Fe-hydrogenase, subunit beta
| Macromolecule | Name: Fe-hydrogenase, subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: hydrogenase (NAD+, ferredoxin) |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) (bacteria) Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) (bacteria)Strain: ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8 |
| Molecular weight | Theoretical: 68.769406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFKNAKEFVQ YANKLKTLRE KKLNGVSIYV CVGTGCTAKG ALKVYSAFEE ELKKRNLLGQ VTLEKIDDDK VTLNRTGCCG RCSSGPLVK IMPYRFFYSN VAPEDVPEIV DRTVLKGEPI ERLFLTDPLT GEKVPRIEDT TLFKNQDFYI MEAIGESECD S IEDYIARS ...String: MFKNAKEFVQ YANKLKTLRE KKLNGVSIYV CVGTGCTAKG ALKVYSAFEE ELKKRNLLGQ VTLEKIDDDK VTLNRTGCCG RCSSGPLVK IMPYRFFYSN VAPEDVPEIV DRTVLKGEPI ERLFLTDPLT GEKVPRIEDT TLFKNQDFYI MEAIGESECD S IEDYIARS GYESLVKALT SMTPEEIIET VKASGLRGRG GGGFPTGLKW EFTRKAQGDI KFVVCNGDEG DPGAFMNRTL LE RDPHLVL EGMIIAGYAV GAQKGYAYIR AEYPFAVKMF KKAIEDARKL GLLGENILGT GFSFDLEVKE GAGAFVCGEE TAL LASIEG KRGMPRPKPP FPAQSGLWGK PTLINNVETY ANIPRILRDG VENYRKRGTE NSPGTKMFSV AGPLKATGII EVEF GTTLR DIIYNICGGF VEGEEFKAVQ IGGPSGACLS EDFIDMPLDY DTLKKADAMV GSGGIVVITK KTCMVEVARF FLDFT KRES CGKCVPCREG TMQAYNILEK FTHGKATYED LKTLEHLSKT IKTASLCGLG KTAPNPILST LKLFREEYIA HIEGEC PSG MCTAFKKYVI NPDICKGCGL CARSCPQNAI TGERGKPYTI DQEKCVKCGL CASKCPFKAI ELV UniProtKB: Bifurcating [FeFe] hydrogenase beta subunit |
-Macromolecule #3: Fe-hydrogenase, subunit gamma
| Macromolecule | Name: Fe-hydrogenase, subunit gamma / type: protein_or_peptide / ID: 3 Details: M initiation codon AS is a linker WSHPQFEK strep tag SGGGGG is a linker ENLYFQ is tev sequence SA is a linker Number of copies: 2 / Enantiomer: LEVO / EC number: hydrogenase (NAD+, ferredoxin) |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) (bacteria) Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) (bacteria)Strain: ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8 |
| Molecular weight | Theoretical: 20.941172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASWSHPQFE KSGGGGGENL YFQGAVLALE RHFEKVEEIL KKYGYKRENL IKILLEIQEI YRYLPEDVIN YVSTAMGIPP AKIYGVATF YAQFSLKPKG KYTIMVCDGT ACHMAGSPEV LKAIEEETGL TPGNVTEDLM FSLDQVGCLG ACALAPVMVI N GEVYGNLT ADKVKEILRK IKEKERESAN V UniProtKB: Bifurcating [FeFe] hydrogenase gamma subunit |
-Macromolecule #4: FE2/S2 (INORGANIC) CLUSTER
| Macromolecule | Name: FE2/S2 (INORGANIC) CLUSTER / type: ligand / ID: 4 / Number of copies: 7 / Formula: FES |
|---|---|
| Molecular weight | Theoretical: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 12 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #6: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 6 / Number of copies: 2 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 346 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||
| Details | Purified by gel filtration |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 4790 / Average exposure time: 6.0 sec. / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 60700 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7p91: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)