+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of Aedes Aegypti Toll5A trimer bound to Spz1C | |||||||||
 Map data Map data | Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mosquito vector biology / Toll receptor / LRR / Cystine knot domain / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationToll binding / central nervous system formation / toll-like receptor signaling pathway / growth factor activity / transmembrane signaling receptor activity / inflammatory response / immune response / innate immune response / extracellular space / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.74 Å | |||||||||
 Authors Authors | Gangloff M / Hardwick SW / Chirgadze DY | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti. Authors: Yoann Saucereau / Thomas H Wilson / Matthew C K Tang / Martin C Moncrieffe / Steven W Hardwick / Dimitri Y Chirgadze / Sandro G Soares / Maria Jose Marcaida / Nicholas J Gay / Monique Gangloff /   Abstract: Aedes aegypti has evolved to become an efficient vector for arboviruses but the mechanisms of host-pathogen tolerance are unknown. Immunoreceptor Toll and its ligand Spaetzle have undergone ...Aedes aegypti has evolved to become an efficient vector for arboviruses but the mechanisms of host-pathogen tolerance are unknown. Immunoreceptor Toll and its ligand Spaetzle have undergone duplication which may allow neofunctionalization and adaptation. Here we present cryo-EM structures and biophysical characterisation of low affinity Toll5A complexes that display transient but specific interactions with Spaetzle1C, forming asymmetric complexes, with only one ligand clearly resolved. Loop structures of Spaetzle1C and Toll5A intercalate, temporarily bridging the receptor C-termini to promote signalling. By contrast unbound receptors form head-to-head homodimers that keep the juxtamembrane regions far apart in an inactive conformation. Interestingly the transcriptional signature of Spaetzle1C differs from other Spaetzle cytokines and controls genes involved in innate immunity, metabolism and tissue regeneration. Taken together our results explain how upregulation of Spaetzle1C in the midgut and Toll5A in the salivary gland shape the concomitant immune response. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11983.map.gz emd_11983.map.gz | 104.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11983-v30.xml emd-11983-v30.xml emd-11983.xml emd-11983.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
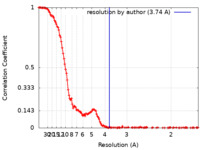
| FSC (resolution estimation) |  emd_11983_fsc.xml emd_11983_fsc.xml | 17.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_11983.png emd_11983.png | 152.1 KB | ||
| Filedesc metadata |  emd-11983.cif.gz emd-11983.cif.gz | 6.6 KB | ||
| Others |  emd_11983_half_map_1.map.gz emd_11983_half_map_1.map.gz emd_11983_half_map_2.map.gz emd_11983_half_map_2.map.gz | 194.2 MB 194.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11983 http://ftp.pdbj.org/pub/emdb/structures/EMD-11983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11983 | HTTPS FTP |
-Validation report
| Summary document |  emd_11983_validation.pdf.gz emd_11983_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11983_full_validation.pdf.gz emd_11983_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_11983_validation.xml.gz emd_11983_validation.xml.gz | 21 KB | Display | |
| Data in CIF |  emd_11983_validation.cif.gz emd_11983_validation.cif.gz | 27.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11983 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11983 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11983 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11983 | HTTPS FTP |
-Related structure data
| Related structure data |  7b1cMC  7b1bC  7b1dC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11983.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11983.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map
| File | emd_11983_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map
| File | emd_11983_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Toll5A-Spz1C complex
| Entire | Name: Toll5A-Spz1C complex |
|---|---|
| Components |
|
-Supramolecule #1: Toll5A-Spz1C complex
| Supramolecule | Name: Toll5A-Spz1C complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: Holodimer complex of Toll5A bound to Spz1C |
|---|---|
| Molecular weight | Theoretical: 230 kDa/nm |
-Supramolecule #2: Toll-like receptor
| Supramolecule | Name: Toll-like receptor / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: AAEL013433-PA
| Supramolecule | Name: AAEL013433-PA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Toll-like receptor
| Macromolecule | Name: Toll-like receptor / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.675773 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TSTKRFTCPE ESEASNCSCE EFPSKTHFYC PDFNPTLYVD VEDRMRVDFK CYDEPHDFKS LPNLAIGSVK LLTVVDCVLD DDRPILESF KFLEVADVRS FVYNNHENGI RYNAKYFEGM EQLENLTLAR GVVSIDRDTF SGFLNLKRLT IEHNKLNLQP G TFEALSNL ...String: TSTKRFTCPE ESEASNCSCE EFPSKTHFYC PDFNPTLYVD VEDRMRVDFK CYDEPHDFKS LPNLAIGSVK LLTVVDCVLD DDRPILESF KFLEVADVRS FVYNNHENGI RYNAKYFEGM EQLENLTLAR GVVSIDRDTF SGFLNLKRLT IEHNKLNLQP G TFEALSNL TYLGLVYNGL NEIQPGLFDG LESLEALSLS YNDIKSLSAG SFNGLSSLRM LNLRVNKIES FDANTFASLK EL SRLEITL NPFVSLPRGL FSENKKLKTL ILTNNRKLVT LPEELLANLK ELTVVNLSHN GVGNLPESLL SGSSGIIELN LGY NRLNSL PEELLSDQPQ LQVLNLDHNQ LESIPDYFLE RNVELQTLYL SHNRLRSLSE KAFTKLKNLK ELHLENNQLQ TIPQ FLFSG TPKLEEIYMQ NNQLALHANS FINEELSIAD NDNTPFQVLQ KLRILHLRNN SISTIFQDWY INNLEMQSLD LSFNK LPGL SYTQLQFQSN ITLNLSNNEI SQVLLIDDLD LQPYQRINVD LNHNPLNCNC NALKFIQLIQ SKAEHGLQFN VDQLRC SEP PNLLDATMDQ LQTKDLLCDF ESADDCPKDC QCAMRLLDHT VIVNCSGRGL TEFPDLPIPS QLHEDFNALE VHVENNR LT KLPNLTKHNE ITQLYARNNS IQNLLPHNIP SKLRIIDLSQ NLLKMIDDST LAQINRSSHL ETIRLSQNQW LCDCPASS F LIFVQQNSRL ISDMSAIRCH PSGKSLDSIT VNELCFEDYT TENLYFQ UniProtKB: TIR domain-containing protein |
-Macromolecule #2: AAEL013433-PA
| Macromolecule | Name: AAEL013433-PA / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.998096 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: GSDTANAPFL CESEQLLIHP KEELSRNNSM VWIVNTKDYK QGVRIEKCLK RQLGKPCNFC DADTECKQLF HYRTLVAVDK VTKKPYKEQ VLLPSCCKCA KILSTGWSHP QFEK UniProtKB: AAEL013433-PA |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK I / Details: Blotting force 0. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.0 sec. / Average electron dose: 51.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 51.1 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)