[English] 日本語
 Yorodumi
Yorodumi- EMDB-11635: Aquifex aeolicus lumazine synthase-derived nucleocapsid variant NC-4 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11635 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Aquifex aeolicus lumazine synthase-derived nucleocapsid variant NC-4 | |||||||||
 Map data Map data | map postprocessed in RELION | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | capsid / design / virus mimic / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology information6,7-dimethyl-8-ribityllumazine synthase / 6,7-dimethyl-8-ribityllumazine synthase activity / riboflavin synthase complex / riboflavin biosynthetic process / RNA polymerase binding / bacterial-type RNA polymerase core enzyme binding / transcription antitermination factor activity, RNA binding / regulation of DNA-templated transcription elongation / transcription antitermination / DNA-templated transcription termination ...6,7-dimethyl-8-ribityllumazine synthase / 6,7-dimethyl-8-ribityllumazine synthase activity / riboflavin synthase complex / riboflavin biosynthetic process / RNA polymerase binding / bacterial-type RNA polymerase core enzyme binding / transcription antitermination factor activity, RNA binding / regulation of DNA-templated transcription elongation / transcription antitermination / DNA-templated transcription termination / RNA stem-loop binding / single-stranded RNA binding / regulation of transcription by RNA polymerase II / DNA binding / RNA binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Escherichia virus lambda / Escherichia virus lambda /   Aquifex aeolicus VF5 (bacteria) Aquifex aeolicus VF5 (bacteria) | |||||||||
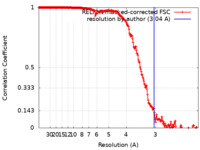
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Tetter S / Hilvert D | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein. Authors: Stephan Tetter / Naohiro Terasaka / Angela Steinauer / Richard J Bingham / Sam Clark / Andrew J P Scott / Nikesh Patel / Marc Leibundgut / Emma Wroblewski / Nenad Ban / Peter G Stockley / ...Authors: Stephan Tetter / Naohiro Terasaka / Angela Steinauer / Richard J Bingham / Sam Clark / Andrew J P Scott / Nikesh Patel / Marc Leibundgut / Emma Wroblewski / Nenad Ban / Peter G Stockley / Reidun Twarock / Donald Hilvert /   Abstract: Viruses are ubiquitous pathogens of global impact. Prompted by the hypothesis that their earliest progenitors recruited host proteins for virion formation, we have used stringent laboratory evolution ...Viruses are ubiquitous pathogens of global impact. Prompted by the hypothesis that their earliest progenitors recruited host proteins for virion formation, we have used stringent laboratory evolution to convert a bacterial enzyme that lacks affinity for nucleic acids into an artificial nucleocapsid that efficiently packages and protects multiple copies of its own encoding messenger RNA. Revealing remarkable convergence on the molecular hallmarks of natural viruses, the accompanying changes reorganized the protein building blocks into an interlaced 240-subunit icosahedral capsid that is impermeable to nucleases, and emergence of a robust RNA stem-loop packaging cassette ensured high encapsidation yields and specificity. In addition to evincing a plausible evolutionary pathway for primordial viruses, these findings highlight practical strategies for developing nonviral carriers for diverse vaccine and delivery applications. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein Authors: Tetter S / Terasaka N / Steinauer A / Bingham RJ / Clark S / Scott AJP / Patel N / Leibundgut M / Wroblewski E / Ban N / Stockley PG / Twarock R / Hilvert D | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11635.map.gz emd_11635.map.gz | 764.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11635-v30.xml emd-11635-v30.xml emd-11635.xml emd-11635.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11635_fsc.xml emd_11635_fsc.xml | 21.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_11635.png emd_11635.png | 118.1 KB | ||
| Masks |  emd_11635_msk_1.map emd_11635_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11635.cif.gz emd-11635.cif.gz | 6.3 KB | ||
| Others |  emd_11635_additional_1.map.gz emd_11635_additional_1.map.gz emd_11635_half_map_1.map.gz emd_11635_half_map_1.map.gz emd_11635_half_map_2.map.gz emd_11635_half_map_2.map.gz | 666.8 MB 669.6 MB 669.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11635 http://ftp.pdbj.org/pub/emdb/structures/EMD-11635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11635 | HTTPS FTP |
-Validation report
| Summary document |  emd_11635_validation.pdf.gz emd_11635_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11635_full_validation.pdf.gz emd_11635_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_11635_validation.xml.gz emd_11635_validation.xml.gz | 27.9 KB | Display | |
| Data in CIF |  emd_11635_validation.cif.gz emd_11635_validation.cif.gz | 37.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11635 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11635 | HTTPS FTP |
-Related structure data
| Related structure data |  7a4jMC  7a4fC  7a4gC  7a4hC  7a4iC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11635.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11635.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map postprocessed in RELION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11635_msk_1.map emd_11635_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: map from 3D autorefinement in RELION
| File | emd_11635_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map from 3D autorefinement in RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_11635_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_11635_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nucleocapsid NC-4
| Entire | Name: Nucleocapsid NC-4 |
|---|---|
| Components |
|
-Supramolecule #1: Nucleocapsid NC-4
| Supramolecule | Name: Nucleocapsid NC-4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Evolved variant of a virus-inspired nucleocapsid design based on the Aquifex aeolicus riboflavin synthase |
|---|
-Supramolecule #2: Nucleocapsid NC-4
| Supramolecule | Name: Nucleocapsid NC-4 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Escherichia virus lambda Escherichia virus lambda |
-Macromolecule #1: Antitermination protein N,6,7-dimethyl-8-ribityllumazine synthase...
| Macromolecule | Name: Antitermination protein N,6,7-dimethyl-8-ribityllumazine synthase,6,7-dimethyl-8-ribityllumazine synthase type: protein_or_peptide / ID: 1 / Number of copies: 240 / Enantiomer: LEVO / EC number: 6,7-dimethyl-8-ribityllumazine synthase |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus VF5 (bacteria) Aquifex aeolicus VF5 (bacteria) |
| Molecular weight | Theoretical: 21.475307 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGNARTRRRE RRAEKQAQWK AANAGAGAGA MATPHFDYNA SVVSKGLANL SLELRKPVSF DIITADTLEQ AIERAGTKHG NKGWEAALS AIEMANLYKS LRGTEHHHHL HGSSIEIYEG KLTAEGLRFG IVASRFNHTL VDRLVEGAID CIVRHGGRGE D ITLVRVPG ...String: MGNARTRRRE RRAEKQAQWK AANAGAGAGA MATPHFDYNA SVVSKGLANL SLELRKPVSF DIITADTLEQ AIERAGTKHG NKGWEAALS AIEMANLYKS LRGTEHHHHL HGSSIEIYEG KLTAEGLRFG IVASRFNHTL VDRLVEGAID CIVRHGGRGE D ITLVRVPG AWEIPVAADE LARKEDIDAV IAFGDLIRG UniProtKB: Antitermination protein N, 6,7-dimethyl-8-ribityllumazine synthase, 6,7-dimethyl-8-ribityllumazine synthase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 12 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 22 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)