[English] 日本語
 Yorodumi
Yorodumi- EMDB-7321: Integrative Structure and Functional Anatomy of a Nuclear Pore Complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7321 | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Integrative Structure and Functional Anatomy of a Nuclear Pore Complex | |||||||||||||||||||||||||||||||||||||||
 Map data Map data | Integrative Structure and Functional Anatomy of a Nuclear Pore Complex | |||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 28.0 Å | |||||||||||||||||||||||||||||||||||||||
 Authors Authors | Kim SJ / Fernandez-Martinez J / Nudelman I / Shi Y / Zhang W / Ludtke SJ / Akey CW / Chait BT / Sali A / Rout MP | |||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, 12 items United States, 12 items
| |||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Integrative structure and functional anatomy of a nuclear pore complex. Authors: Seung Joong Kim / Javier Fernandez-Martinez / Ilona Nudelman / Yi Shi / Wenzhu Zhang / Barak Raveh / Thurston Herricks / Brian D Slaughter / Joanna A Hogan / Paula Upla / Ilan E Chemmama / ...Authors: Seung Joong Kim / Javier Fernandez-Martinez / Ilona Nudelman / Yi Shi / Wenzhu Zhang / Barak Raveh / Thurston Herricks / Brian D Slaughter / Joanna A Hogan / Paula Upla / Ilan E Chemmama / Riccardo Pellarin / Ignacia Echeverria / Manjunatha Shivaraju / Azraa S Chaudhury / Junjie Wang / Rosemary Williams / Jay R Unruh / Charles H Greenberg / Erica Y Jacobs / Zhiheng Yu / M Jason de la Cruz / Roxana Mironska / David L Stokes / John D Aitchison / Martin F Jarrold / Jennifer L Gerton / Steven J Ludtke / Christopher W Akey / Brian T Chait / Andrej Sali / Michael P Rout /  Abstract: Nuclear pore complexes play central roles as gatekeepers of RNA and protein transport between the cytoplasm and nucleoplasm. However, their large size and dynamic nature have impeded a full ...Nuclear pore complexes play central roles as gatekeepers of RNA and protein transport between the cytoplasm and nucleoplasm. However, their large size and dynamic nature have impeded a full structural and functional elucidation. Here we determined the structure of the entire 552-protein nuclear pore complex of the yeast Saccharomyces cerevisiae at sub-nanometre precision by satisfying a wide range of data relating to the molecular arrangement of its constituents. The nuclear pore complex incorporates sturdy diagonal columns and connector cables attached to these columns, imbuing the structure with strength and flexibility. These cables also tie together all other elements of the nuclear pore complex, including membrane-interacting regions, outer rings and RNA-processing platforms. Inwardly directed anchors create a high density of transport factor-docking Phe-Gly repeats in the central channel, organized into distinct functional units. This integrative structure enables us to rationalize the architecture, transport mechanism and evolutionary origins of the nuclear pore complex. | |||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7321.map.gz emd_7321.map.gz | 18.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7321-v30.xml emd-7321-v30.xml emd-7321.xml emd-7321.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7321.png emd_7321.png | 271.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7321 http://ftp.pdbj.org/pub/emdb/structures/EMD-7321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7321 | HTTPS FTP |
-Validation report
| Summary document |  emd_7321_validation.pdf.gz emd_7321_validation.pdf.gz | 80.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7321_full_validation.pdf.gz emd_7321_full_validation.pdf.gz | 79.3 KB | Display | |
| Data in XML |  emd_7321_validation.xml.gz emd_7321_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7321 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7321 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7321 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7321 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10155 (Title: Cryo-electron tomography of the yeast NPC / Data size: 127.1 EMPIAR-10155 (Title: Cryo-electron tomography of the yeast NPC / Data size: 127.1 Data #1: Unaligned K2 DED image tilt series of yeast NPC and associated files [micrographs - single frame])  EMPIAR-10162 (Title: Integrative structure and functional anatomy of a nuclear pore complex EMPIAR-10162 (Title: Integrative structure and functional anatomy of a nuclear pore complexData size: 11.8 Data #1: Raw micrographs of Nic96 complex [micrographs - single frame] Data #2: Sc Nic96 complex class averages [class averages]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7321.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7321.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Integrative Structure and Functional Anatomy of a Nuclear Pore Complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
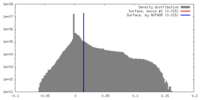
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae NPC (nuclear pore complex)
| Entire | Name: Saccharomyces cerevisiae NPC (nuclear pore complex) |
|---|---|
| Components |
|
-Supramolecule #1: Saccharomyces cerevisiae NPC (nuclear pore complex)
| Supramolecule | Name: Saccharomyces cerevisiae NPC (nuclear pore complex) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Affinity-purified isolated whole NPCs |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 87 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 Details: A medium thick carbon film was used to support the NPCs over the holes. Before use, the grids were glow discharged in air, floated on 5 uL sample drops for 45 minutes and then washed by ...Details: A medium thick carbon film was used to support the NPCs over the holes. Before use, the grids were glow discharged in air, floated on 5 uL sample drops for 45 minutes and then washed by serial transfer on 4 x 20 micro-litre drops of sample buffer without glycerol. | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK III Details: Buffer on the grid was removed by blotting from the bottom with a tool that held a filter paper wedge, using access through the left-hand port. Then 2 micro-litre of freezing buffer was ...Details: Buffer on the grid was removed by blotting from the bottom with a tool that held a filter paper wedge, using access through the left-hand port. Then 2 micro-litre of freezing buffer was added to the grid from the right-hand port and the grid was plunge frozen in liquid ethane after blotting.. | ||||||||||||||||||||||||
| Details | Sample isolated in one affinity step - pullout from frozen yeast cell grindate. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Titan-Krios equipped with a spherical aberration (Cs) corrector Chromatic aberration corrector: none / Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: 1 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | The electron gun was an XFEG. A total of 253 tilt series were collected in steps between -60, 0 and 60 degrees in increments of 2.5 - 4 degrees for different tilt series. While the full tilt range was used for tomogram reconstruction, in the final subtomogram averaging step only data up to +/-45 degrees tilt from each sub-volume were included in the final average. A defocus range of -4.6 to -7.5 microns was used. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Average electron dose: 3.0 e/Å2 Details: The dose target for each tilt series was 90-100 electrons/angstrom squared and followed a cosine alpha dose curve with a flux of 20 electrons/pixel/second, and a dose of 3.5 ...Details: The dose target for each tilt series was 90-100 electrons/angstrom squared and followed a cosine alpha dose curve with a flux of 20 electrons/pixel/second, and a dose of 3.5 electrons/angstrom squared for the zero tilt image. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 9434 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal defocus max: 7.5 µm / Nominal defocus min: 4.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C8 (8 fold cyclic) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 28.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: EMAN2 (ver. 2.1) Details: The final map has been fully CTF corrected and filtered based on the estimated local resolution. Number subtomograms used: 1864 |
|---|---|
| Extraction | Number tomograms: 121 / Number images used: 6416 / Method: manual selection / Software - Name: EMAN2 (ver. 2.12) / Software - details: e2spt_boxer.py Details: A low pass filter of 100 Angstroms was applied to binned 3X SIRT tomograms prior to particle picking. Final unbinned sub-volumes of 300 x 300 x 300 were extracted from the original tomograms. |
| CTF correction | Software - Name: EMAN2 (ver. 2.1) Details: Phase flipping of tilted images with etomo in IMOD running in batchtomo mode. The final reconstruction used 1,864 (of the 6,416 initial) particles. Theoretical CTF curves for the mean ...Details: Phase flipping of tilted images with etomo in IMOD running in batchtomo mode. The final reconstruction used 1,864 (of the 6,416 initial) particles. Theoretical CTF curves for the mean defocus values present in the tomograms were averaged assuming 10% amplitude contrast. The reciprocal of this curve was then applied as a filter to the final uncorrected map. |
| Final angle assignment | Type: OTHER / Software - Name: EMAN2 Software - details: e2spt_classaverage.py in theEMAN2 single particle tomography package Details: Iterative 3D alignment of sub-volumes to a reference volume, using the C8 symmetry of the NPC. |
-Atomic model buiding 1
| Details | The integrative structure modeling protocol was scripted using the Python Modeling Interface (PMI) package, version 4d97507, a library for modeling macromolecular complexes based on our open-source Integrative Modeling Platform (IMP) package, version 2.6 (https://integrativemodeling.org) |
|---|---|
| Refinement | Protocol: OTHER |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)