[English] 日本語
 Yorodumi
Yorodumi- EMDB-52158: Nuclear ring of the nuclear pore complex from Nup133-deficient mo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nuclear ring of the nuclear pore complex from Nup133-deficient mouse embryonic stem cell | |||||||||
 Map data Map data | Nuclear ring of the nuclear pore complex from Nup133-deficient mouse embryonic stem cell | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / nuclear transport / nuclear envelope / TRANSPORT PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 33.4 Å | |||||||||
 Authors Authors | Taniguchi R / Beck M | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Cell Biol / Year: 2025 Journal: Nat Cell Biol / Year: 2025Title: Nuclear pores safeguard the integrity of the nuclear envelope. Authors: Reiya Taniguchi / Clarisse Orniacki / Jan Philipp Kreysing / Vojtech Zila / Christian E Zimmerli / Stefanie Böhm / Beata Turoňová / Hans-Georg Kräusslich / Valérie Doye / Martin Beck /      Abstract: Nuclear pore complexes (NPCs) mediate nucleocytoplasmic exchange, which is essential for eukaryotes. Mutations in the central scaffolding components of NPCs are associated with genetic diseases, but ...Nuclear pore complexes (NPCs) mediate nucleocytoplasmic exchange, which is essential for eukaryotes. Mutations in the central scaffolding components of NPCs are associated with genetic diseases, but how they manifest only in specific tissues remains unclear. This is exemplified in Nup133-deficient mouse embryonic stem cells, which grow normally during pluripotency, but differentiate poorly into neurons. Here, using an innovative in situ structural biology approach, we show that Nup133 mouse embryonic stem cells have heterogeneous NPCs with non-canonical symmetries and missing subunits. During neuronal differentiation, Nup133-deficient NPCs frequently disintegrate, resulting in abnormally large nuclear envelope openings. We propose that the elasticity of the NPC scaffold has a protective function for the nuclear envelope and that its perturbation becomes critical under conditions that impose an increased mechanical load onto nuclei. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_52158.map.gz emd_52158.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-52158-v30.xml emd-52158-v30.xml emd-52158.xml emd-52158.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_52158_fsc.xml emd_52158_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_52158.png emd_52158.png | 134.4 KB | ||
| Masks |  emd_52158_msk_1.map emd_52158_msk_1.map | 3.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-52158.cif.gz emd-52158.cif.gz | 4.2 KB | ||
| Others |  emd_52158_half_map_1.map.gz emd_52158_half_map_1.map.gz emd_52158_half_map_2.map.gz emd_52158_half_map_2.map.gz | 3.5 MB 3.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-52158 http://ftp.pdbj.org/pub/emdb/structures/EMD-52158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52158 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_52158.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_52158.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nuclear ring of the nuclear pore complex from Nup133-deficient mouse embryonic stem cell | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 10.728 Å | ||||||||||||||||||||||||||||||||||||
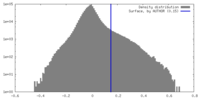
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_52158_msk_1.map emd_52158_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
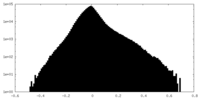
| Density Histograms |
-Half map: Half map of the nuclear ring of the...
| File | emd_52158_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of the nuclear ring of the nuclear pore complex from Nup133-deficient mouse embryonic stem cell | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of the nuclear ring of the...
| File | emd_52158_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of the nuclear ring of the nuclear pore complex from Nup133-deficient mouse embryonic stem cell | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Inner ring of the nuclear pore complex from wild-type mouse embry...
| Entire | Name: Inner ring of the nuclear pore complex from wild-type mouse embryonic stem cell |
|---|---|
| Components |
|
-Supramolecule #1: Inner ring of the nuclear pore complex from wild-type mouse embry...
| Supramolecule | Name: Inner ring of the nuclear pore complex from wild-type mouse embryonic stem cell type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 300 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 6 / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)