[English] 日本語
 Yorodumi
Yorodumi- EMDB-44174: Cryo-electron tomographic investigation of native hippocampal glu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomographic investigation of native hippocampal glutamatergic synapses - Tomogram 1 | |||||||||
 Map data Map data | Tomogram of glutamatergic synapse | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Synapse AMPA Neurotransmission / MEMBRANE PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Matsui A / Spangler CJ / Elferich J / Gouaux E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Cryo-electron tomographic investigation of native hippocampal glutamatergic synapses. Authors: Aya Matsui / Cathy Spangler / Johannes Elferich / Momoko Shiozaki / Nikki Jean / Xiaowei Zhao / Maozhen Qin / Haining Zhong / Zhiheng Yu / Eric Gouaux /  Abstract: Chemical synapses are the major sites of communication between neurons in the nervous system and mediate either excitatory or inhibitory signaling. At excitatory synapses, glutamate is the primary ...Chemical synapses are the major sites of communication between neurons in the nervous system and mediate either excitatory or inhibitory signaling. At excitatory synapses, glutamate is the primary neurotransmitter and upon release from presynaptic vesicles, is detected by postsynaptic glutamate receptors, which include ionotropic AMPA and NMDA receptors. Here, we have developed methods to identify glutamatergic synapses in brain tissue slices, label AMPA receptors with small gold nanoparticles (AuNPs), and prepare lamella for cryo-electron tomography studies. The targeted imaging of glutamatergic synapses in the lamella is facilitated by fluorescent pre- and postsynaptic signatures, and the subsequent tomograms allow for the identification of key features of chemical synapses, including synaptic vesicles, the synaptic cleft, and AuNP-labeled AMPA receptors. These methods pave the way for imaging brain regions at high resolution, using unstained, unfixed samples preserved under near-native conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44174.map.gz emd_44174.map.gz | 1.1 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44174-v30.xml emd-44174-v30.xml emd-44174.xml emd-44174.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44174.png emd_44174.png | 143.2 KB | ||
| Filedesc metadata |  emd-44174.cif.gz emd-44174.cif.gz | 4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44174 http://ftp.pdbj.org/pub/emdb/structures/EMD-44174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44174 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44174.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44174.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomogram of glutamatergic synapse | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 10 Å | ||||||||||||||||||||||||||||||||
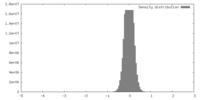
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Glutamatergic synapse from mouse hippocampus tissue
| Entire | Name: Glutamatergic synapse from mouse hippocampus tissue |
|---|---|
| Components |
|
-Supramolecule #1: Glutamatergic synapse from mouse hippocampus tissue
| Supramolecule | Name: Glutamatergic synapse from mouse hippocampus tissue / type: tissue / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 20 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: at 15 mA |
| Vitrification | Cryogen name: NITROGEN |
| Details | Dissected hippocampus tissue from vGlut1-mScarlet/PSD95-EGFP mouse line |
| High pressure freezing | Instrument: OTHER Details: The value given for _em_high_pressure_freezing.instrument is Leica Ice. This is not in a list of allowed values {'EMS-002 RAPID IMMERSION FREEZER', 'LEICA EM PACT', 'LEICA EM HPM100', 'LEICA ...Details: The value given for _em_high_pressure_freezing.instrument is Leica Ice. This is not in a list of allowed values {'EMS-002 RAPID IMMERSION FREEZER', 'LEICA EM PACT', 'LEICA EM HPM100', 'LEICA EM PACT2', 'BAL-TEC HPM 010', 'OTHER'} so OTHER is written into the XML file. |
| Cryo protectant | 20% dextran; 5% sucrose |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 / Focused ion beam - Current: 0.03 / Focused ion beam - Duration: 120 / Focused ion beam - Temperature: 83 K / Focused ion beam - Initial thickness: 40000 / Focused ion beam - Final thickness: 200 Focused ion beam - Details: The value given for _em_focused_ion_beam.instrument is TFS Aquilos 2. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 4.55 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 2.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name: RELION (ver. 4.0.1) / Number images used: 33 |
|---|
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)