+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of Caulobacter crescentus Tad pilus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tight adhesion pilus / Flp family type IVb pilin / CELL ADHESION | |||||||||
| Function / homology | Flp/Fap pilin component / Flp/Fap pilin component / membrane / PilA Function and homology information Function and homology information | |||||||||
| Biological species |  Caulobacter vibrioides (bacteria) Caulobacter vibrioides (bacteria) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Sonani RR / Sanchez JC / Baumgardt JK / Wright ER / Egelman EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Tad and toxin-coregulated pilus structures reveal unexpected diversity in bacterial type IV pili. Authors: Ravi R Sonani / Juan Carlos Sanchez / Joseph K Baumgardt / Shivani Kundra / Elizabeth R Wright / Lisa Craig / Edward H Egelman /   Abstract: Type IV pili (T4P) are ubiquitous in both bacteria and archaea. They are polymers of the major pilin protein, which has an extended and protruding N-terminal helix, α1, and a globular C-terminal ...Type IV pili (T4P) are ubiquitous in both bacteria and archaea. They are polymers of the major pilin protein, which has an extended and protruding N-terminal helix, α1, and a globular C-terminal domain. Cryo-EM structures have revealed key differences between the bacterial and archaeal T4P in their C-terminal domain structure and in the packing and continuity of α1. This segment forms a continuous α-helix in archaeal T4P but is partially melted in all published bacterial T4P structures due to a conserved helix breaking proline at position 22. The tad (tight adhesion) T4P are found in both bacteria and archaea and are thought to have been acquired by bacteria through horizontal transfer from archaea. Tad pilins are unique among the T4 pilins, being only 40 to 60 residues in length and entirely lacking a C-terminal domain. They also lack the Pro22 found in all high-resolution bacterial T4P structures. We show using cryo-EM that the bacterial tad pilus from is composed of continuous helical subunits that, like the archaeal pilins, lack the melted portion seen in other bacterial T4P and share the packing arrangement of the archaeal T4P. We further show that a bacterial T4P, the toxin coregulated pilus, which lacks Pro22 but is not in the tad family, has a continuous N-terminal α-helix, yet its α1 s are arranged similar to those in other bacterial T4P. Our results highlight the role of Pro22 in helix melting and support an evolutionary relationship between tad and archaeal T4P. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41815.map.gz emd_41815.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41815-v30.xml emd-41815-v30.xml emd-41815.xml emd-41815.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41815_fsc.xml emd_41815_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_41815.png emd_41815.png | 40.7 KB | ||
| Filedesc metadata |  emd-41815.cif.gz emd-41815.cif.gz | 4.7 KB | ||
| Others |  emd_41815_half_map_1.map.gz emd_41815_half_map_1.map.gz emd_41815_half_map_2.map.gz emd_41815_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41815 http://ftp.pdbj.org/pub/emdb/structures/EMD-41815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41815 | HTTPS FTP |
-Validation report
| Summary document |  emd_41815_validation.pdf.gz emd_41815_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41815_full_validation.pdf.gz emd_41815_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_41815_validation.xml.gz emd_41815_validation.xml.gz | 18.9 KB | Display | |
| Data in CIF |  emd_41815_validation.cif.gz emd_41815_validation.cif.gz | 23.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41815 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41815 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41815 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41815 | HTTPS FTP |
-Related structure data
| Related structure data |  8u1kMC  8uhfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41815.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41815.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41815_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
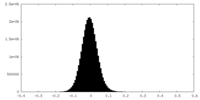
| Density Histograms |
-Half map: #1
| File | emd_41815_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
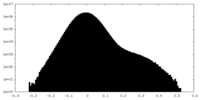
| Density Histograms |
- Sample components
Sample components
-Entire : Tad pilus filament
| Entire | Name: Tad pilus filament |
|---|---|
| Components |
|
-Supramolecule #1: Tad pilus filament
| Supramolecule | Name: Tad pilus filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Caulobacter vibrioides (bacteria) Caulobacter vibrioides (bacteria) |
-Macromolecule #1: PilA
| Macromolecule | Name: PilA / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Caulobacter vibrioides (bacteria) Caulobacter vibrioides (bacteria) |
| Molecular weight | Theoretical: 4.365073 KDa |
| Sequence | String: ATAIEYGLIV ALIAVVIVTA VTTLGTNLRT AFTKAGAAVS TAAGT UniProtKB: PilA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)