+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
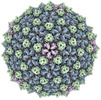
| Title | Mycobacterium phage Adjutor | |||||||||
 Map data Map data | Sharpened map of ewald sphere corrected postprocess. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | T=7 / HK97 / Tailed bacteriophage / Capsid / VIRUS | |||||||||
| Function / homology | Uncharacterized protein / Uncharacterized protein / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Mycobacterium phage Adjutor (virus) Mycobacterium phage Adjutor (virus) | |||||||||
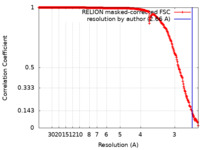
| Method | single particle reconstruction / cryo EM / Resolution: 2.66 Å | |||||||||
 Authors Authors | Podgorski JM / White SJ | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Stabilization mechanism accommodating genome length variation in evolutionarily related viral capsids. Authors: Jennifer M Podgorski / Joshua Podgorski / Lawrence Abad / Deborah Jacobs-Sera / Krista G Freeman / Colin Brown / Graham F Hatfull / Antoni Luque / Simon J White /  Abstract: Tailed bacteriophages are one of the most numerous and diverse group of viruses. They store their genome at quasi-crystalline densities in capsids built from multiple copies of proteins adopting the ...Tailed bacteriophages are one of the most numerous and diverse group of viruses. They store their genome at quasi-crystalline densities in capsids built from multiple copies of proteins adopting the HK97-fold. The high density of the genome exerts an internal pressure, requiring a maturation process that reinforces their capsids. However, it is unclear how capsid stabilization strategies have adapted to accommodate the evolution of larger genomes in this virus group. Here we characterize a capsid reinforcement mechanism in two evolutionary-related actinobacteriophages that modifies the length of a stabilization protein to accommodate a larger genome while maintaining the same capsid size. We use cryo-EM to reveal that capsids contain split hexamers of HK97-fold proteins with a stabilization protein in the chasm. The observation of split hexamers in mature capsids is unprecedented, so we rationalize this result mathematically, discovering that icosahedral capsids can be formed by all split or skewed hexamers as long as their T-number is not a multiple of three. Our results suggest that analogous stabilization mechanisms can be present in other icosahedral capsids, and they provide a strategy for engineering capsids accommodating larger DNA cargoes as gene delivery systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40271.map.gz emd_40271.map.gz | 1.7 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40271-v30.xml emd-40271-v30.xml emd-40271.xml emd-40271.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40271_fsc.xml emd_40271_fsc.xml | 27.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40271.png emd_40271.png | 293 KB | ||
| Masks |  emd_40271_msk_1.map emd_40271_msk_1.map | 1.9 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-40271.cif.gz emd-40271.cif.gz | 6.4 KB | ||
| Others |  emd_40271_additional_1.map.gz emd_40271_additional_1.map.gz emd_40271_half_map_1.map.gz emd_40271_half_map_1.map.gz emd_40271_half_map_2.map.gz emd_40271_half_map_2.map.gz | 1.8 GB 1.5 GB 1.5 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40271 http://ftp.pdbj.org/pub/emdb/structures/EMD-40271 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40271 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40271 | HTTPS FTP |
-Related structure data
| Related structure data |  8sajMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40271.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40271.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of ewald sphere corrected postprocess. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40271_msk_1.map emd_40271_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Ewald sphere corrected map.
| File | emd_40271_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ewald sphere corrected map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of ewald sphere corrected map.
| File | emd_40271_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of ewald sphere corrected map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of ewald sphere corrected map.
| File | emd_40271_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of ewald sphere corrected map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterium phage Adjutor
| Entire | Name:  Mycobacterium phage Adjutor (virus) Mycobacterium phage Adjutor (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium phage Adjutor
| Supramolecule | Name: Mycobacterium phage Adjutor / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 528321 / Sci species name: Mycobacterium phage Adjutor / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Virus shell | Shell ID: 1 / Diameter: 750.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium phage Adjutor (virus) Mycobacterium phage Adjutor (virus) |
| Molecular weight | Theoretical: 45.525645 KDa |
| Sequence | String: MKTATEGRVM RESFLEIIAV VGLSGHDNQG GYNTAGDIKY KTADGVSYDS LWNLFSNVTD EWNKHKSKMV QLMTFPVTNQ TEKVPRIGQ FGFEKASEFG VPESKRTELS FYQLAYDFED YDLAFRYTWK FLRDAPSSQI KAYHNQALQA DAKLIHRKVM E AIFDNRER ...String: MKTATEGRVM RESFLEIIAV VGLSGHDNQG GYNTAGDIKY KTADGVSYDS LWNLFSNVTD EWNKHKSKMV QLMTFPVTNQ TEKVPRIGQ FGFEKASEFG VPESKRTELS FYQLAYDFED YDLAFRYTWK FLRDAPSSQI KAYHNQALQA DAKLIHRKVM E AIFDNRER EADIEGLPYK VYPLYNGDNM IPPEYNGTTF STGHNHYLVS GGTKIDSADV EMAADHIREH GYTEENGTQL IA FAHKAEI QEVRRFRFGQ TNNNSAVANY DFVQSQGESP LYLPNADGLL GKQPQSMWKG LRVKGSYDDV LWIEEPTMPA GYV LFLATG GTLAQQNLVG LREHEDAAWR GLRQIPGNQT RYPLIDSFYQ RSFGTGIRQR GGAVVLQIKA SGTYDIPTKW TNGG GFE UniProtKB: Major capsid protein |
-Macromolecule #2: gp_16 (Minor Capsid Protein)
| Macromolecule | Name: gp_16 (Minor Capsid Protein) / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium phage Adjutor (virus) Mycobacterium phage Adjutor (virus) |
| Molecular weight | Theoretical: 13.766352 KDa |
| Sequence | String: MARYDKYNPY GGGFRAPLAA DWTDADAGKL YAVGINNVGA VVKGAGQSGV AGVLVLTKGA KAGSIVDVMK FGEVVEFGPT SGTPGTDFG AAGTAYYADT STGAINSTSG EAKVKVGHTV GAQRLIVAVA DGVVDPSPAA UniProtKB: Uncharacterized protein |
-Macromolecule #3: HNH endonuclease
| Macromolecule | Name: HNH endonuclease / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium phage Adjutor (virus) Mycobacterium phage Adjutor (virus) |
| Molecular weight | Theoretical: 6.198103 KDa |
| Sequence | String: MAKGVKKLPK RKGTNPIPRD KWNSDDIARR QLEQDQKLHL TTKGPHTGTN DSFK UniProtKB: Uncharacterized protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number real images: 6664 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Amino acid sequence built into the map for a single major capsid protein and refined with Phenix. Model then used for rest of asymmetric unit and refined with Phenix. Final step involved using Isolde. |
|---|---|
| Refinement | Protocol: AB INITIO MODEL |
| Output model |  PDB-8saj: |
 Movie
Movie Controller
Controller



 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)