+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Porcine uroplakin complex | ||||||||||||||||||
 Map data Map data | Uroplakin complex | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Urothelium / asymmetric unit membrane / urinary bladder / urinary tract infection / lipid raft / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationapical plasma membrane urothelial plaque / urinary bladder development / urea transport / water transport / sodium ion homeostasis / potassium ion homeostasis / epithelial cell differentiation / kidney development / cell morphogenesis / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
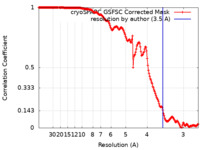
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||||||||
 Authors Authors | Oda T / Yanagisawa H / Kikkawa M | ||||||||||||||||||
| Funding support |  Japan, 5 items Japan, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Cryo-EM elucidates the uroplakin complex structure within liquid-crystalline lipids in the porcine urothelial membrane. Authors: Haruaki Yanagisawa / Yoshihiro Kita / Toshiyuki Oda / Masahide Kikkawa /  Abstract: The urothelium, a distinct epithelial tissue lining the urinary tract, serves as an essential component in preserving urinary tract integrity and thwarting infections. The asymmetric unit membrane ...The urothelium, a distinct epithelial tissue lining the urinary tract, serves as an essential component in preserving urinary tract integrity and thwarting infections. The asymmetric unit membrane (AUM), primarily composed of the uroplakin complex, constitutes a critical permeability barrier in fulfilling this role. However, the molecular architectures of both the AUM and the uroplakin complex have remained enigmatic due to the paucity of high-resolution structural data. In this study, we utilized cryo-electron microscopy to elucidate the three-dimensional structure of the uroplakin complex within the porcine AUM. While the global resolution achieved was 3.5 Å, we acknowledge that due to orientation bias, the resolution in the vertical direction was determined to be 6.3 Å. Our findings unveiled that the uroplakin complexes are situated within hexagonally arranged crystalline lipid membrane domains, rich in hexosylceramides. Moreover, our research rectifies a misconception in a previous model by confirming the existence of a domain initially believed to be absent, and pinpointing the accurate location of a crucial Escherichia coli binding site implicated in urinary tract infections. These discoveries offer valuable insights into the molecular underpinnings governing the permeability barrier function of the urothelium and the orchestrated lipid phase formation within the plasma membrane. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36340.map.gz emd_36340.map.gz | 28.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36340-v30.xml emd-36340-v30.xml emd-36340.xml emd-36340.xml | 24.8 KB 24.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36340_fsc.xml emd_36340_fsc.xml | 16.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_36340.png emd_36340.png | 93.2 KB | ||
| Masks |  emd_36340_msk_1.map emd_36340_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36340.cif.gz emd-36340.cif.gz | 7.4 KB | ||
| Others |  emd_36340_half_map_1.map.gz emd_36340_half_map_1.map.gz emd_36340_half_map_2.map.gz emd_36340_half_map_2.map.gz | 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36340 http://ftp.pdbj.org/pub/emdb/structures/EMD-36340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36340 | HTTPS FTP |
-Related structure data
| Related structure data |  8jj5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36340.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36340.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Uroplakin complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
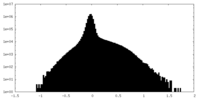
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36340_msk_1.map emd_36340_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
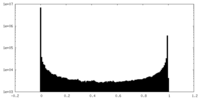
| Density Histograms |
-Half map: half map A
| File | emd_36340_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
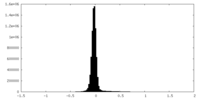
| Density Histograms |
-Half map: half map B
| File | emd_36340_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
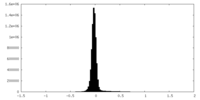
| Density Histograms |
- Sample components
Sample components
-Entire : Uroplakin complex
| Entire | Name: Uroplakin complex |
|---|---|
| Components |
|
-Supramolecule #1: Uroplakin complex
| Supramolecule | Name: Uroplakin complex / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Asymmetric unit membrane isolated by sarkosyl extraction |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tetraspanin
| Macromolecule | Name: Tetraspanin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.747469 KDa |
| Sequence | String: MASAAAEGEK GSPVVVGLLV VGNIIILLSG LALFAETIWV TADQYRVYPL MGVSGKDDVF AGAWIAIFCG FSFFVVASFG VGAALCRRR SMILTYLVLM LIVYIFECAS CITSYTHRDY MVSNPSLITK QMLTFYSADS DQGRELTRLW DRVMIEQECC G TSGPMDWV ...String: MASAAAEGEK GSPVVVGLLV VGNIIILLSG LALFAETIWV TADQYRVYPL MGVSGKDDVF AGAWIAIFCG FSFFVVASFG VGAALCRRR SMILTYLVLM LIVYIFECAS CITSYTHRDY MVSNPSLITK QMLTFYSADS DQGRELTRLW DRVMIEQECC G TSGPMDWV NFTSAFRAST PEVVFPWPPL CCRRTGNFIP VNEEGCRLGH LDYLFTKGCF EHIGHAIDSY TWGISWFGFA IL MWTLPVM LIAMYFYTTL UniProtKB: Tetraspanin |
-Macromolecule #2: Uroplakin 2
| Macromolecule | Name: Uroplakin 2 / type: protein_or_peptide / ID: 2 / Details: NCBI Reference Sequence: NP_999177.1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.299869 KDa |
| Sequence | String: MASPLPVRTL PLILILLAVL APGASDFNIS SLSGPLSPAL TESLLVALPP CHLTGGNATL MVRRANDSKV VKSSFMVPPC RGRRELVSV VDSGSGFTVT RLSAYQVTNL VPGTKYYISY LVTKGASTES SREIPMSTLP RRKAEAIGLG MAPTGGMVVI Q VLLSVAMF LLVVGFITAL ALGARK |
-Macromolecule #3: Uroplakin 3A
| Macromolecule | Name: Uroplakin 3A / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.339234 KDa |
| Sequence | String: MPLLWALLAL GCLQLGSGVN LQPQLASVTF ATNNPTLTTV ALEKPLCMFD SSAALHGTYE VYLYVLVDSA SSRNASVQDS TKTPLSSTP QETEGGRTGP YKAAAFDLAP CSDLPSLDAV RDVSQASEIL NAYLVRVGIN GTCLSDPNFR GLCNPPLSAA T EYRFKYVL ...String: MPLLWALLAL GCLQLGSGVN LQPQLASVTF ATNNPTLTTV ALEKPLCMFD SSAALHGTYE VYLYVLVDSA SSRNASVQDS TKTPLSSTP QETEGGRTGP YKAAAFDLAP CSDLPSLDAV RDVSQASEIL NAYLVRVGIN GTCLSDPNFR GLCNPPLSAA T EYRFKYVL VNISTGLVQD QTLWSDPVCT NQLTPYSAID TWPGRRSGGM IVITSILGSL PFFLLVGFAG AIVLSLMDMG GA DGEMTHD SQITQEAVPK GTSEPSYTSV NRGPPLDRAE VYASKLQD UniProtKB: Uroplakin 3A |
-Macromolecule #4: UPK1B
| Macromolecule | Name: UPK1B / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.857492 KDa |
| Sequence | String: MAKDDSTVRC FQSLLVFGNV IIGMCGIALT AECIFFVSDQ YSLYPLLEAT DNDDIYGAAW IGIFVGICLF CLSVLGIVGI MKSNRKILL VYFILMFIVY GFEVASCITA ATQRDFFTPN LFLKQMLERY QNNSPPSNDD KWKNNGVTKT WDRLMLQDYC C GVNGPSDW ...String: MAKDDSTVRC FQSLLVFGNV IIGMCGIALT AECIFFVSDQ YSLYPLLEAT DNDDIYGAAW IGIFVGICLF CLSVLGIVGI MKSNRKILL VYFILMFIVY GFEVASCITA ATQRDFFTPN LFLKQMLERY QNNSPPSNDD KWKNNGVTKT WDRLMLQDYC C GVNGPSDW QKYTSAFRTE NNDADYPWPR QCCVMNKLKE PLNLEACKLG VPGYYHNQGC YELISGPMNR HAWGVAWFGF AI LCWTFWV LLGTMFYWSR IEY UniProtKB: Uroplakin-1b |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER/RHODIUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8jj5: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)