+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lloviu cuevavirus nucleoprotein-RNA complex | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | nucleoprotein / VIRAL PROTEIN | |||||||||||||||||||||
| Function / homology | Ebola nucleoprotein / Ebola nucleoprotein / viral RNA genome packaging / helical viral capsid / viral nucleocapsid / host cell cytoplasm / ribonucleoprotein complex / RNA binding / Nucleoprotein Function and homology information Function and homology information | |||||||||||||||||||||
| Biological species |  Lloviu cuevavirus / synthetic construct (others) Lloviu cuevavirus / synthetic construct (others) | |||||||||||||||||||||
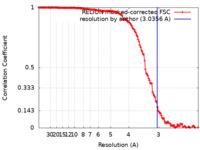
| Method | single particle reconstruction / cryo EM / Resolution: 3.0356 Å | |||||||||||||||||||||
 Authors Authors | Hu SF / Fujita-Fujiharu Y / Sugita Y / Wendt L / Muramoto Y / Nakano M / Hoenen T / Noda T | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: PNAS Nexus / Year: 2023 Journal: PNAS Nexus / Year: 2023Title: Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus. Authors: Shangfan Hu / Yoko Fujita-Fujiharu / Yukihiko Sugita / Lisa Wendt / Yukiko Muramoto / Masahiro Nakano / Thomas Hoenen / Takeshi Noda /   Abstract: Lloviu virus (LLOV) is a novel filovirus detected in Schreiber's bats in Europe. The isolation of the infectious LLOV from bats has raised public health concerns. However, the virological and ...Lloviu virus (LLOV) is a novel filovirus detected in Schreiber's bats in Europe. The isolation of the infectious LLOV from bats has raised public health concerns. However, the virological and molecular characteristics of LLOV remain largely unknown. The nucleoprotein (NP) of LLOV encapsidates the viral genomic RNA to form a helical NP-RNA complex, which acts as a scaffold for nucleocapsid formation and de novo viral RNA synthesis. In this study, using single-particle cryoelectron microscopy, we determined two structures of the LLOV NP-RNA helical complex, comprising a full-length and a C-terminally truncated NP. The two helical structures were identical, demonstrating that the N-terminal region determines the helical arrangement of the NP. The LLOV NP-RNA protomers displayed a structure similar to that in the Ebola and Marburg virus, but the spatial arrangements in the helix differed. Structure-based mutational analysis identified amino acids involved in the helical assembly and viral RNA synthesis. These structures advance our understanding of the filovirus nucleocapsid formation and provide a structural basis for the development of antifiloviral therapeutics. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34016.map.gz emd_34016.map.gz | 228.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34016-v30.xml emd-34016-v30.xml emd-34016.xml emd-34016.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34016_fsc.xml emd_34016_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_34016.png emd_34016.png | 234.4 KB | ||
| Filedesc metadata |  emd-34016.cif.gz emd-34016.cif.gz | 7 KB | ||
| Others |  emd_34016_half_map_1.map.gz emd_34016_half_map_1.map.gz emd_34016_half_map_2.map.gz emd_34016_half_map_2.map.gz | 194 MB 193.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34016 http://ftp.pdbj.org/pub/emdb/structures/EMD-34016 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34016 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34016 | HTTPS FTP |
-Related structure data
| Related structure data |  7ypwMC  7yr8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34016.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34016.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.13 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34016_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
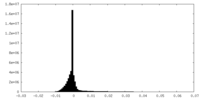
| Density Histograms |
-Half map: #1
| File | emd_34016_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Lloviu cuevavirus nucleoprotein RNA complex
| Entire | Name: Lloviu cuevavirus nucleoprotein RNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Lloviu cuevavirus nucleoprotein RNA complex
| Supramolecule | Name: Lloviu cuevavirus nucleoprotein RNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Lloviu cuevavirus Lloviu cuevavirus |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lloviu cuevavirus / Strain: isolate Bat/Spain/Asturias-Bat86/2003 Lloviu cuevavirus / Strain: isolate Bat/Spain/Asturias-Bat86/2003 |
| Molecular weight | Theoretical: 84.553094 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNRYLGHGTR TSRENTNLSE LHGILSLGLN VDHTIVRKKS IPLFEIGNSD QVCNWIIQII EAGVDLQEVA DSFLTMLCVN HAYQGDPNL FLESPAAHYL KGHGIHFEIQ HRDNVDHITD LLGVGSRDKS LRKTLSALEF EPGGTTTAGM FLSFASLFLP K LVVGERAC ...String: MNRYLGHGTR TSRENTNLSE LHGILSLGLN VDHTIVRKKS IPLFEIGNSD QVCNWIIQII EAGVDLQEVA DSFLTMLCVN HAYQGDPNL FLESPAAHYL KGHGIHFEIQ HRDNVDHITD LLGVGSRDKS LRKTLSALEF EPGGTTTAGM FLSFASLFLP K LVVGERAC LEKVQRQIQI HAEQGLIQYP TQWQSVGHMM VVFRLIRVNF VLKFLLVHQG MHMMAGHDAN DAIIANSISQ TR FSGLLIV KTVLEHILQK TEAGVQLHPL ARTSKVKGEL LAFKSALEAL ASHREYAPFA RLLNLSGVNN LEHGLYPQLS AIA LGVATA HGSTLAGVNV SEQYQQLREA ATEAEKQLQQ HSEMRELETL GLDEQERKIL ATFHSRKNEI NIQQTSSILA IRKE RLRKL TEALTEEKNK NALDDEDESE EDDWSPENRG IRSNKGSTKE PSSYTASRTE EDRNNYNSKD DHLSGKEQMS TQQES GADD LDLFDLDDDG DTNSQDPNNR QKQSDTQQTQ ESSDRSDYSR RPAYDWPPGD RPHTTQATDE HTDLLNKDHR RNQVKP GRR GNDPRTLPLI SFDDNEGEIL DDKSDLPAPD THSDPDTEES EEEHPDEELL PPAPKYSTKT SEQEPGDWKQ PTSPLST IP EEEGGHEANN DNSESDLIDQ MYQHIFKTEG AYAAINYYYK TTGRPVTFTS NNNHDYTFPQ DTEGLFPPWE GKENQKVA E ILTNSLHETG QEWADMSAKE RYLFLINN UniProtKB: Nucleoprotein |
-Macromolecule #2: RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.792037 KDa |
| Sequence | String: UUUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Apply 1.25 micro litters of sample from each side of the grid and blot for 7 seconds before plugging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector with two hexapole elements |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 3122 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 59000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)