+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of PldA-PA3488 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PldA / PA3488 / Complex / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationphospholipid catabolic process / D-type glycerophospholipase activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) | |||||||||
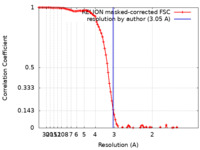
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Zhao L / Yang XY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural insights into PA3488-mediated inactivation of Pseudomonas aeruginosa PldA. Authors: Xiaoyun Yang / Zongqiang Li / Liang Zhao / Zhun She / Zengqiang Gao / Sen-Fang Sui / Yuhui Dong / Yanhua Li /  Abstract: PldA, a phospholipase D (PLD) effector, catalyzes hydrolysis of the phosphodiester bonds of glycerophospholipids-the main component of cell membranes-and assists the invasion of the opportunistic ...PldA, a phospholipase D (PLD) effector, catalyzes hydrolysis of the phosphodiester bonds of glycerophospholipids-the main component of cell membranes-and assists the invasion of the opportunistic pathogen Pseudomonas aeruginosa. As a cognate immunity protein, PA3488 can inhibit the activity of PldA to avoid self-toxicity. However, the precise inhibitory mechanism remains elusive. We determine the crystal structures of full-length and truncated PldA and the cryogenic electron microscopy structure of the PldA-PA3488 complex. Structural analysis reveals that there are different intermediates of PldA between the "open" and "closed" states of the catalytic pocket, accompanied by significant conformational changes in the "lid" region and the peripheral helical domain. Through structure-based mutational analysis, we identify the key residues responsible for the enzymatic activity of PldA. Together, these data provide an insight into the molecular mechanisms of PldA invasion and its neutralization by PA3488, aiding future design of PLD-targeted inhibitors and drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32438.map.gz emd_32438.map.gz | 85.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32438-v30.xml emd-32438-v30.xml emd-32438.xml emd-32438.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32438_fsc.xml emd_32438_fsc.xml | 9.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_32438.png emd_32438.png | 50.4 KB | ||
| Filedesc metadata |  emd-32438.cif.gz emd-32438.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32438 http://ftp.pdbj.org/pub/emdb/structures/EMD-32438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32438 | HTTPS FTP |
-Related structure data
| Related structure data |  7wdkMC  7v53C  7v55C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32438.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32438.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.668 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : The structure of PldA-PA3488 complex
| Entire | Name: The structure of PldA-PA3488 complex |
|---|---|
| Components |
|
-Supramolecule #1: The structure of PldA-PA3488 complex
| Supramolecule | Name: The structure of PldA-PA3488 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Phospholipase D
| Macromolecule | Name: Phospholipase D / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 122.478062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLQKKPYNGL HEKELNQINQ QDGSPCVAIS APGCFIKGSN LFSEKRAGNR VRFFTTGRDY FSDLASALDS ASSSIFITGW QVNYDVLLD GRRSLWQCLR QALERSPALK VYVMPWLSPS GSLGTYDFET MLAVFQLNAG LEGGARAFCT PAIQQSDMQG L GVAFSHHQ ...String: MLQKKPYNGL HEKELNQINQ QDGSPCVAIS APGCFIKGSN LFSEKRAGNR VRFFTTGRDY FSDLASALDS ASSSIFITGW QVNYDVLLD GRRSLWQCLR QALERSPALK VYVMPWLSPS GSLGTYDFET MLAVFQLNAG LEGGARAFCT PAIQQSDMQG L GVAFSHHQ KSVVIDNRIG YVGGIDLAYG RRDDNDFSLD ASGRRGNDAY NPGLPHLGWM AEDEHVSSMG LMMATLFDLS RP LASLTLH APTLRLSPFP HIAASDEPLL SIPLAPSRAR ALNGAAYLSD LFRSPMLPSL QWLGRAYNSS KEGLDEGFER LDA LRRQMV ASSIRAIANL IADNLDALPI EPELERRLRA WLEELRTAAL NLPEALRIKS LLLINQWMSE TELGQVLTLI SGKG FEDIP QNLSGKAGEL AGSLFWTLHR LMQARAGGHQ QPYRYLDEAP QPLASPDNAR LAADQPRMPW QDVHCRIEGP SVYDL ARNF IDRWNGQQAY LAKTPALQDT ALVRSALEAV MKWLNSLAAA AGLENYLDEK RNLRLELDPP TPCWINAPEQ LPQEPE VRR GGMTVQVLRS AAARMLEQEQ AGRLGAGVNL PLQVGVSTEG VQSNCKDAML LAISGAQQFI YIENQFFQSE FGKEGEV FK DLPLSGPMAS LRDVGSLRRD FVVRIRLEEA LEQRDLWLLD WAEVEKIAQE PGTEARQFLK SMLAMWGVNA QGWLTHKL G EAQHGLLNEI GEALARRIER AIQREHPFHV YLVLPVHPEG ALNVPNIMHQ VHLTQQSLVF GEQSLVKRIQ RQMALKALE GKSDPAQARE IIERKDARGR PVYEQQDWSR YLTLLNLRTW AVLGGRVVTE QIYVHSKLLI ADDRVAILGS ANINDRSLQG ERDSELAVM VRDSEPLTVR LDGKNDAIVG KAIHQLRVNL WKKHFGLSQG PGGFVKPASE LSAYLSIPAA QEAWEAIQTL A KENTRAYE RTFNFIPQNI SQTQLQLTPE PPKGFEDGFP ASIWPTWAYR KPGELRAGGQ LMEPMPYQEI FWRSSNLTSV KT FPPPNGV SGFITALPTS WTRGERNDSG LNLSILAHQD SRSLPTQVAM NGDSSAQGKH RT UniProtKB: Phospholipase D |
-Macromolecule #2: Tli4_C domain-containing protein
| Macromolecule | Name: Tli4_C domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.088637 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKRVLMGLIL LSSSNITWAE APSSKYQECL GRMTFEIPEE MEWATYDASR VWQISKGGGH NFTAEVTAVG DNGSYDYDSM IFYVSEKVD KNEFHNASNY IKGTAEIYQD HLRENIKLDK KAISTLQKNK SEEKSIERIK KGIAEMEAKI PLAKIYEHDL G IPDSHILG ...String: MKRVLMGLIL LSSSNITWAE APSSKYQECL GRMTFEIPEE MEWATYDASR VWQISKGGGH NFTAEVTAVG DNGSYDYDSM IFYVSEKVD KNEFHNASNY IKGTAEIYQD HLRENIKLDK KAISTLQKNK SEEKSIERIK KGIAEMEAKI PLAKIYEHDL G IPDSHILG SKNIPFHVLL WRNQRVYYFT FSKPTENSAQ RIKDLIARFR TRELYEVPNE PGICFPYGFI ADDGKTAYEL KN SLRFTRT PNVIFSLLTA SANDPWQTRP TSGLYDSDFR PGYDRQKWKK SALLDSLHIG KRLAAFEGWR LDPRPDSGER ERA WFGLAH TGGTLDPLVA IQVQTFQKGT DDLTDYTPPP EEVLPRLKAL SQSIEQRLAR UniProtKB: Tle cognate immunity protein 4 C-terminal domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)