[English] 日本語
 Yorodumi
Yorodumi- EMDB-32118: Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / RNA BINDING PROTEIN-RNA COMPLEX | |||||||||
| Function / homology | Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |  Planctomycetes bacterium (bacteria) / synthetic construct (others) Planctomycetes bacterium (bacteria) / synthetic construct (others) | |||||||||
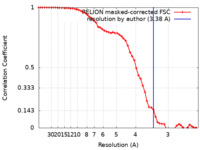
| Method | single particle reconstruction / cryo EM / Resolution: 3.38 Å | |||||||||
 Authors Authors | Nakagawa R / Soumya K / Han A / Takeda NS / Tomita A / Hirano H / Kusakizako T / Tomohiro N / Yamashita K / Feng Z ...Nakagawa R / Soumya K / Han A / Takeda NS / Tomita A / Hirano H / Kusakizako T / Tomohiro N / Yamashita K / Feng Z / Nishimasu H / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structure and engineering of the minimal type VI CRISPR-Cas13bt3. Authors: Ryoya Nakagawa / Soumya Kannan / Han Altae-Tran / Satoru N Takeda / Atsuhiro Tomita / Hisato Hirano / Tsukasa Kusakizako / Tomohiro Nishizawa / Keitaro Yamashita / Feng Zhang / Hiroshi ...Authors: Ryoya Nakagawa / Soumya Kannan / Han Altae-Tran / Satoru N Takeda / Atsuhiro Tomita / Hisato Hirano / Tsukasa Kusakizako / Tomohiro Nishizawa / Keitaro Yamashita / Feng Zhang / Hiroshi Nishimasu / Osamu Nureki /    Abstract: Type VI CRISPR-Cas13 effector enzymes catalyze RNA-guided RNA cleavage and have been harnessed for various technologies, such as RNA detection, targeting, and editing. Recent studies identified ...Type VI CRISPR-Cas13 effector enzymes catalyze RNA-guided RNA cleavage and have been harnessed for various technologies, such as RNA detection, targeting, and editing. Recent studies identified Cas13bt3 (also known as Cas13X.1) as a miniature Cas13 enzyme, which can be used for knockdown and editing of target transcripts in mammalian cells. However, the action mechanism of the compact Cas13bt3 remains unknown. Here, we report the structures of the Cas13bt3-guide RNA complex and the Cas13bt3-guide RNA-target RNA complex. The structures revealed how Cas13bt3 recognizes the guide RNA and its target RNA and provided insights into the activation mechanism of Cas13bt3, which is distinct from those of the other Cas13a/d enzymes. Furthermore, we rationally engineered enhanced Cas13bt3 variants and ultracompact RNA base editors. Overall, this study improves our mechanistic understanding of the CRISPR-Cas13 enzymes and paves the way for the development of efficient Cas13-mediated transcriptome modulation technologies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32118.map.gz emd_32118.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32118-v30.xml emd-32118-v30.xml emd-32118.xml emd-32118.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32118_fsc.xml emd_32118_fsc.xml | 5.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_32118.png emd_32118.png | 108.1 KB | ||
| Masks |  emd_32118_msk_1.map emd_32118_msk_1.map | 9.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32118.cif.gz emd-32118.cif.gz | 6 KB | ||
| Others |  emd_32118_half_map_1.map.gz emd_32118_half_map_1.map.gz emd_32118_half_map_2.map.gz emd_32118_half_map_2.map.gz | 7.9 MB 7.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32118 http://ftp.pdbj.org/pub/emdb/structures/EMD-32118 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32118 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32118 | HTTPS FTP |
-Related structure data
| Related structure data |  7vtnMC  7vtiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32118.map.gz / Format: CCP4 / Size: 9.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32118.map.gz / Format: CCP4 / Size: 9.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21733 Å | ||||||||||||||||||||||||||||||||||||
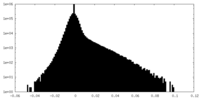
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_32118_msk_1.map emd_32118_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
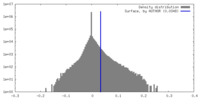
| Density Histograms |
-Half map: #2
| File | emd_32118_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
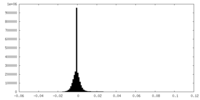
| Density Histograms |
-Half map: #1
| File | emd_32118_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
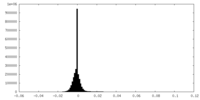
| Density Histograms |
- Sample components
Sample components
-Entire : Cas13bt3-crRNA-target RNA
| Entire | Name: Cas13bt3-crRNA-target RNA |
|---|---|
| Components |
|
-Supramolecule #1: Cas13bt3-crRNA-target RNA
| Supramolecule | Name: Cas13bt3-crRNA-target RNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: crRNA-target RNA
| Supramolecule | Name: crRNA-target RNA / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Planctomycetes bacterium (bacteria) / Synthetically produced: Yes Planctomycetes bacterium (bacteria) / Synthetically produced: Yes |
-Supramolecule #3: Cas13bt3
| Supramolecule | Name: Cas13bt3 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|
-Macromolecule #1: Cas13bt3
| Macromolecule | Name: Cas13bt3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Planctomycetes bacterium (bacteria) Planctomycetes bacterium (bacteria) |
| Molecular weight | Theoretical: 90.498789 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGMAQVSKQT SKKRELSIDE YQGARKWCFT IAFNKALVNR DKNDGLFVES LLRHEKYSKH DWYDEDTRAL IKCSTQAANA KAEALANYF SAYRHSPGCL TFTAEDELRT IMERAYERAI FECRRRETEV IIEFPSLFEG DRITTAGVVF FVSFFVERRV L DRLYGAVS ...String: GGMAQVSKQT SKKRELSIDE YQGARKWCFT IAFNKALVNR DKNDGLFVES LLRHEKYSKH DWYDEDTRAL IKCSTQAANA KAEALANYF SAYRHSPGCL TFTAEDELRT IMERAYERAI FECRRRETEV IIEFPSLFEG DRITTAGVVF FVSFFVERRV L DRLYGAVS GLKKNEGQYK LTRKALSMYC LKDSRFTKAW DKRVLLFRDI LAQLGRIPAE AYEYYHGEQG DKKRANDNEG TN PKRHKDK FIEFALHYLE AQHSEICFGR RHIVREEAGA GDEHKKHRTK GKVVVDFSKK DEDQSYYISK NNVIVRIDKN AGP RSYRMG LNELKYLVLL SLQGKGDDAI AKLYRYRQHV ENILDVVKVT DKDNHVFLPR FVLEQHGIGR KAFKQRIDGR VKHV RGVWE KKKAATNEMT LHEKARDILQ YVNENCTRSF NPGEYNRLLV CLVGKDVENF QAGLKRLQLA ERIDGRVYSI FAQTS TINE MHQVVCDQIL NRLCRIGDQK LYDYVGLGKK DEIDYKQKVA WFKEHISIRR GFLRKKFWYD SKKGFAKLVE EHLESG GGQ RDVGLDKKYY HIDAIGRFEG ANPALYETLA RDRLCLMMAQ YFLGSVRKEL GNKIVWSNDS IELPVEGSVG NEKSIVF SV SDYGKLYVLD DAEFLGRICE YFMPHEKGKI RYHTVYEKGF RAYNDLQKKC VEAVLAFEEK VVKAKKMSEK EGAHYIDF R EILAQTMCKE AEKTAVNKVA RAFFAHHLKF VIDEFGLFSD VMKKYGIEKE WKFPVK UniProtKB: Uncharacterized protein |
-Macromolecule #2: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 19.603619 KDa |
| Sequence | String: GCUUGGCAAC CAUUCAAAUA UGUAUGCUGG AGCAGCCCCC GAUUUGUGGG GUGAUUACAG C |
-Macromolecule #3: target RNA
| Macromolecule | Name: target RNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.984773 KDa |
| Sequence | String: AUACAUAUUU GAAUGGUUGC CAAGC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 48.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)