[English] 日本語
 Yorodumi
Yorodumi- EMDB-32043: Complex structure of Clostridioides difficile enzymatic component... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Translocation / Oligomer / Unfoldase / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein homooligomerization / transferase activity / extracellular region / metal ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) | |||||||||
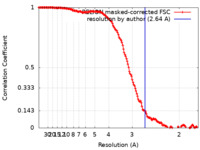
| Method | single particle reconstruction / cryo EM / Resolution: 2.64 Å | |||||||||
 Authors Authors | Yamada T / Kawamoto A / Yoshida T / Sato Y / Kato T / Tsuge H | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile. Authors: Akihiro Kawamoto / Tomohito Yamada / Toru Yoshida / Yusui Sato / Takayuki Kato / Hideaki Tsuge /  Abstract: Some bacteria express a binary toxin translocation system, consisting of an enzymatic subunit and translocation pore, that delivers enzymes into host cells through endocytosis. The most clinically ...Some bacteria express a binary toxin translocation system, consisting of an enzymatic subunit and translocation pore, that delivers enzymes into host cells through endocytosis. The most clinically important bacterium with such a system is Clostridioides difficile (formerly Clostridium). The CDTa and CDTb proteins from its system represent important therapeutic targets. CDTb has been proposed to be a di-heptamer, but its physiological heptameric structure has not yet been reported. Here, we report the cryo-EM structure of CDTa bound to the CDTb-pore, which reveals that CDTa binding induces partial unfolding and tilting of the first CDTa α-helix. In the CDTb-pore, an NSS-loop exists in 'in' and 'out' conformations, suggesting its involvement in substrate translocation. Finally, 3D variability analysis revealed CDTa movements from a folded to an unfolded state. These dynamic structural information provide insights into drug design against hypervirulent C. difficile strains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32043.map.gz emd_32043.map.gz | 24.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32043-v30.xml emd-32043-v30.xml emd-32043.xml emd-32043.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32043_fsc.xml emd_32043_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_32043.png emd_32043.png | 125.2 KB | ||
| Filedesc metadata |  emd-32043.cif.gz emd-32043.cif.gz | 6.4 KB | ||
| Others |  emd_32043_additional_1.map.gz emd_32043_additional_1.map.gz | 245.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32043 http://ftp.pdbj.org/pub/emdb/structures/EMD-32043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32043 | HTTPS FTP |
-Related structure data
| Related structure data |  7vnnMC  7vnjC  7yvqC  7yvsC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32043.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32043.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_32043_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CDTa-bound CDTb-pore (long)
| Entire | Name: CDTa-bound CDTb-pore (long) |
|---|---|
| Components |
|
-Supramolecule #1: CDTa-bound CDTb-pore (long)
| Supramolecule | Name: CDTa-bound CDTb-pore (long) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
-Supramolecule #2: CDTb-pore
| Supramolecule | Name: CDTb-pore / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
-Supramolecule #4: CdtA
| Supramolecule | Name: CdtA / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: ADP-ribosylating binary toxin binding subunit CdtB
| Macromolecule | Name: ADP-ribosylating binary toxin binding subunit CdtB / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
| Molecular weight | Theoretical: 75.657141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NNNFFDPKLM SDWEDEDLDT DNDNIPDSYE RNGYTIKDLI AVKWEDSFAE QGYKKYVSNY LESNTAGDPY TDYEKASGSF DKAIKTEAR DPLVAAYPIV GVGMEKLIIS TNEHASTDQG KTVSRATTNS KTESNTAGVS VNVGYQNGFT ANVTTNYSHT T DNSTAVQD ...String: NNNFFDPKLM SDWEDEDLDT DNDNIPDSYE RNGYTIKDLI AVKWEDSFAE QGYKKYVSNY LESNTAGDPY TDYEKASGSF DKAIKTEAR DPLVAAYPIV GVGMEKLIIS TNEHASTDQG KTVSRATTNS KTESNTAGVS VNVGYQNGFT ANVTTNYSHT T DNSTAVQD SNGESWNTGL SINKGESAYI NANVRYYNTG TAPMYKVTPT TNLVLDGDTL STIKAQENQI GNNLSPGDTY PK KGLSPLA LNTMDQFSSR LIPINYDQLK KLDAGKQIKL ETTQVSGNFG TKNSSGQIVT EGNSWSDYIS QIDSISASII LDT ENESYE RRVTAKNLQD PEDKTPELTI GEAIEKAFGA TKKDGLLYFN DIPIDESCVE LIFDDNTANK IKDSLKTLSD KKIY NVKLE RGMNILIKTP TYFTNFDDYN NYPSTWSNVN TTNQDGLQGS ANKLNGETKI KIPMSELKPY KRYVFSGYSK DPLTS NSII VKIKAKEEKT DYLVPEQGYT KFSYEFETTE KDSSNIEITL IGSGTTYLDN LSITELNSTP EILDEPEVKI PTDQEI MDA HKIYFADLNF NPSTGNTYIN GMYFAPTQTN KEALDYIQKY RVEATLQYSG FKDIGTKDKE MRNYLGDPNQ PKTNYVN LR SYFTGGENIM TYKKLRIYAI TPDDRELLVL SVD UniProtKB: ADP-ribosyltransferase binding component |
-Macromolecule #2: CdtA
| Macromolecule | Name: CdtA / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
| Molecular weight | Theoretical: 49.420469 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: APIERPEDFL KDKEKAKEWE RKEAERIEQK LERSEKEALE SYKKDSVEIS KYSQTRNYFY DYQIEANSRE KEYKELRNAI SKNKIDKPM YVYYFESPEK FAFNKVIRTE NQNEISLEKF NEFKETIQNK LFKQDGFKDI SLYEPGKGDE KPTPLLMHLK L PRNTGMLP ...String: APIERPEDFL KDKEKAKEWE RKEAERIEQK LERSEKEALE SYKKDSVEIS KYSQTRNYFY DYQIEANSRE KEYKELRNAI SKNKIDKPM YVYYFESPEK FAFNKVIRTE NQNEISLEKF NEFKETIQNK LFKQDGFKDI SLYEPGKGDE KPTPLLMHLK L PRNTGMLP YTNTNNVSTL IEQGYSIKID KIVRIVIDGK HYIKAEASVV SSLDFKDDVS KGDSWGKANY NDWSNKLTPN EL ADVNDYM RGGYTAINNY LISNGPVNNP NPELDSKITN IENALKREPI PTNLTVYRRS GPQEFGLTLT SPEYDFNKLE NID AFKSKW EGQALSYPNF ISTSIGSVNM SAFAKRKIVL RITIPKGSPG AYLSAIPGYA GEYEVLLNHG SKFKINKIDS YKDG TITKL IVDATLIPEN LYFQGLEHHH HHH UniProtKB: CdtA |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 21 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.58 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 11284 / Average exposure time: 3.36 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)