[English] 日本語
 Yorodumi
Yorodumi- EMDB-31081: Subtomogram average of a meiotic triple helix from a pachytene S.... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of a meiotic triple helix from a pachytene S. cerevisiae cell. | ||||||||||||
 Map data Map data | NKY611 meiotic triple helix | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 33.0 Å | ||||||||||||
 Authors Authors | Ma OX / Chong W / Lee JKE / Cai S / Siebert CA / Howe A / Zhang P / Shi J / Surana U / Gan L | ||||||||||||
| Funding support |  Singapore, 3 items Singapore, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS One / Year: 2022 Journal: PLoS One / Year: 2022Title: Cryo-ET detects bundled triple helices but not ladders in meiotic budding yeast. Authors: Olivia X Ma / Wen Guan Chong / Joy K E Lee / Shujun Cai / C Alistair Siebert / Andrew Howe / Peijun Zhang / Jian Shi / Uttam Surana / Lu Gan /   Abstract: In meiosis, cells undergo two sequential rounds of cell division, termed meiosis I and meiosis II. Textbook models of the meiosis I substage called pachytene show that nuclei have conspicuous 100-nm- ...In meiosis, cells undergo two sequential rounds of cell division, termed meiosis I and meiosis II. Textbook models of the meiosis I substage called pachytene show that nuclei have conspicuous 100-nm-wide, ladder-like synaptonemal complexes and ordered chromatin loops. It remains unknown if these cells have any other large, meiosis-related intranuclear structures. Here we present cryo-ET analysis of frozen-hydrated budding yeast cells before, during, and after pachytene. We found no cryo-ET densities that resemble dense ladder-like structures or ordered chromatin loops. Instead, we found large numbers of 12-nm-wide triple-helices that pack into ordered bundles. These structures, herein called meiotic triple helices (MTHs), are present in meiotic cells, but not in interphase cells. MTHs are enriched in the nucleus but not enriched in the cytoplasm. Bundles of MTHs form at the same timeframe as synaptonemal complexes (SCs) in wild-type cells and in mutant cells that are unable to form SCs. These results suggest that in yeast, SCs coexist with previously unreported large, ordered assemblies. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31081.map.gz emd_31081.map.gz | 732.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31081-v30.xml emd-31081-v30.xml emd-31081.xml emd-31081.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31081.png emd_31081.png | 55.5 KB | ||
| Others |  emd_31081_half_map_1.map.gz emd_31081_half_map_1.map.gz emd_31081_half_map_2.map.gz emd_31081_half_map_2.map.gz | 732.7 KB 733.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31081 http://ftp.pdbj.org/pub/emdb/structures/EMD-31081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31081 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31081.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31081.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NKY611 meiotic triple helix | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.61 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 2
| File | emd_31081_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
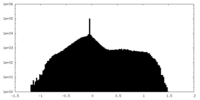
| Density Histograms |
-Half map: Half map 1
| File | emd_31081_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
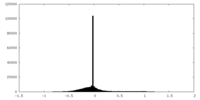
| Density Histograms |
- Sample components
Sample components
-Entire : Subtomogram average of a triple helix from pachytene S. cerevisia...
| Entire | Name: Subtomogram average of a triple helix from pachytene S. cerevisiae cells. |
|---|---|
| Components |
|
-Supramolecule #1: Subtomogram average of a triple helix from pachytene S. cerevisia...
| Supramolecule | Name: Subtomogram average of a triple helix from pachytene S. cerevisiae cells. type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: Sporulation medium (2% potassium acetate) |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: NITROGEN / Instrument: FEI VITROBOT MARK IV Details: The Vitrobot's cup was used to condense the ethane cryogen. Crimped copper tubes containing cell paste were held ~ 3 cm above the surface of the liquid ethane, then dropped.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Number grids imaged: 1 / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 30400 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 18000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 33.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: RELION (ver. 3.0.7) / Number subtomograms used: 841 |
|---|---|
| Extraction | Number tomograms: 13 / Number images used: 841 |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)