[English] 日本語
 Yorodumi
Yorodumi- EMDB-28585: Cryo-EM structure of a delivery complex containing the SspB adapt... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a delivery complex containing the SspB adaptor, an ssrA-tagged substrate, and the AAA+ ClpXP protease | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ protease / ClpXP / SspB adaptor / HYDROLASE / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationHslUV protease complex / endopeptidase Clp / endopeptidase Clp complex / positive regulation of programmed cell death / response to temperature stimulus / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / proteasomal protein catabolic process / : / serine-type peptidase activity ...HslUV protease complex / endopeptidase Clp / endopeptidase Clp complex / positive regulation of programmed cell death / response to temperature stimulus / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / proteasomal protein catabolic process / : / serine-type peptidase activity / bioluminescence / generation of precursor metabolites and energy / ATP-dependent protein folding chaperone / response to radiation / positive regulation of protein catabolic process / unfolded protein binding / peptidase activity / response to heat / ATPase binding / protein dimerization activity / ribosome / serine-type endopeptidase activity / cell division / ATP hydrolysis activity / zinc ion binding / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
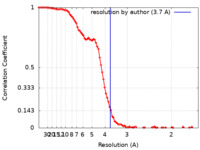
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Ghanbarpour A / Fei X / Davis JH / Sauer RT | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery. Authors: Alireza Ghanbarpour / Xue Fei / Tania A Baker / Joseph H Davis / Robert T Sauer /  Abstract: Energy-dependent protein degradation by the AAA+ ClpXP protease helps maintain protein homeostasis in bacteria and eukaryotic organelles of bacterial origin. In and many other proteobacteria, the ...Energy-dependent protein degradation by the AAA+ ClpXP protease helps maintain protein homeostasis in bacteria and eukaryotic organelles of bacterial origin. In and many other proteobacteria, the SspB adaptor assists ClpXP in degrading ssrA-tagged polypeptides produced as a consequence of tmRNA-mediated ribosome rescue. By tethering these incomplete ssrA-tagged proteins to ClpXP, SspB facilitates their efficient degradation at low substrate concentrations. How this process occurs structurally is unknown. Here, we present a cryo-EM structure of the SspB adaptor bound to a GFP-ssrA substrate and to ClpXP. This structure provides evidence for simultaneous contacts of SspB and ClpX with the ssrA tag within the tethering complex, allowing direct substrate handoff concomitant with the initiation of substrate translocation. Furthermore, our structure reveals that binding of the substrate·adaptor complex induces unexpected conformational changes within the spiral structure of the AAA+ ClpX hexamer and its interaction with the ClpP tetradecamer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28585.map.gz emd_28585.map.gz | 58 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28585-v30.xml emd-28585-v30.xml emd-28585.xml emd-28585.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28585_fsc.xml emd_28585_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_28585.png emd_28585.png | 45.9 KB | ||
| Masks |  emd_28585_msk_1.map emd_28585_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28585.cif.gz emd-28585.cif.gz | 6.7 KB | ||
| Others |  emd_28585_half_map_1.map.gz emd_28585_half_map_1.map.gz emd_28585_half_map_2.map.gz emd_28585_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28585 http://ftp.pdbj.org/pub/emdb/structures/EMD-28585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28585 | HTTPS FTP |
-Related structure data
| Related structure data |  8et3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28585.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28585.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28585_msk_1.map emd_28585_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28585_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_28585_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ClpXP AAA protease complex bound with SspB and GFP-ssrA
| Entire | Name: ClpXP AAA protease complex bound with SspB and GFP-ssrA |
|---|---|
| Components |
|
-Supramolecule #1: ClpXP AAA protease complex bound with SspB and GFP-ssrA
| Supramolecule | Name: ClpXP AAA protease complex bound with SspB and GFP-ssrA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-dependent Clp protease ATP-binding subunit ClpX
| Macromolecule | Name: ATP-dependent Clp protease ATP-binding subunit ClpX / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.414848 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDKRKDGSG KLLYCSFCGK SQHEVRKLIA GPSVYICDEC VDLCNDIIRE EIKEVAPHRE RSALPTPHEI RNHLDDYVIG QEQAKKVLA VAVYNHYKRL RNGDTSNGVE LGKSNILLIG PTGSGKTLLA ETLARLLDVP FTMADATTLT EAGYVGEDVE N IIQKLLQK ...String: MTDKRKDGSG KLLYCSFCGK SQHEVRKLIA GPSVYICDEC VDLCNDIIRE EIKEVAPHRE RSALPTPHEI RNHLDDYVIG QEQAKKVLA VAVYNHYKRL RNGDTSNGVE LGKSNILLIG PTGSGKTLLA ETLARLLDVP FTMADATTLT EAGYVGEDVE N IIQKLLQK CDYDVQKAQR GIVYIDEIDK ISRKSDNPSI TRDVSGEGVQ QALLKLIEGT VAAVPPQGGR KHPQQEFLQV DT SKILFIC GGAFAGLDKV ISHRVETGSG IGFGATVKAK SDKASEGELL AQVEPEDLIK FGLIPEFIGR LPVVATLNEL SEE ALIQIL KEPKNALTKQ YQALFNLEGV DLEFRDEALD AIAKKAMARK TGARGLRSIV EAALLDTMYD LPSMEDVEKV VIDE SVIDG QSEPLLIYGK PEAQQASGE UniProtKB: ATP-dependent Clp protease ATP-binding subunit ClpX |
-Macromolecule #2: ATP-dependent Clp protease proteolytic subunit
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.468869 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LVPMVIEQTS RGERSFDIYS RLLKERVIFL TGQVEDHMAN LIVAQMLFLE AENPEKDIYL YINSPGGVIT AGMSIYDTMQ FIKPDVSTI CMGQAASMGA FLLTAGAKGK RFCLPNSRVM IHQPLGGYQG QATDIEIHAR EILKVKGRMN ELMALHTGQS L EQIERDTE ...String: LVPMVIEQTS RGERSFDIYS RLLKERVIFL TGQVEDHMAN LIVAQMLFLE AENPEKDIYL YINSPGGVIT AGMSIYDTMQ FIKPDVSTI CMGQAASMGA FLLTAGAKGK RFCLPNSRVM IHQPLGGYQG QATDIEIHAR EILKVKGRMN ELMALHTGQS L EQIERDTE RDRFLSAPEA VEYGLVDSIL THRNENLYFQ SLEHHHHHH UniProtKB: ATP-dependent Clp protease proteolytic subunit |
-Macromolecule #3: Green fluorescent protein
| Macromolecule | Name: Green fluorescent protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.855428 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH DYDIPTTENL YFQGSRKGEE LFTGVVPILV ELDGDVNGHK FSVSGEGEGD ATYGKLTLKF ICTTGKLPVP WPTLVTTFG YGVQCFARYP DHMKQHDFFK SAMPEGYVQE RTIFFKDDGN YKTRAEVKFE GDTLVNRIEL KGIDFKEDGN I LGHKLEYN ...String: MGSSHHHHHH DYDIPTTENL YFQGSRKGEE LFTGVVPILV ELDGDVNGHK FSVSGEGEGD ATYGKLTLKF ICTTGKLPVP WPTLVTTFG YGVQCFARYP DHMKQHDFFK SAMPEGYVQE RTIFFKDDGN YKTRAEVKFE GDTLVNRIEL KGIDFKEDGN I LGHKLEYN YNSHNVYIMA DKQKNGIKVN FKIRHNIEDG SVQLADHYQQ NTPIGDGPVL LPDNHYLSTQ SALSKDPNEK RD HMVLLEF VTAAGITHGM DELYKAANDE NYALAA UniProtKB: Green fluorescent protein |
-Macromolecule #4: Stringent starvation protein B
| Macromolecule | Name: Stringent starvation protein B / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.279352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDLSQLTPRR PYLLRAFYEW LLDNQLTPHL VVDVTLPGVQ VPMEYARDGQ IVLNIAPRAV GNLELANDEV RFNARFGGIP RQVSVPLAA VLAIYARENG AGTMFEPEAA YDEDTSIMND EEASADNETV MSVIDGDKPD HDDDTHPDDE PPQPPRGGRP A LRVVK UniProtKB: Stringent starvation protein B |
-Macromolecule #5: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 4 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 75.98 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)