[English] 日本語
 Yorodumi
Yorodumi- EMDB-2834: Cryo-EM structure of TMV reconstructed from micrographs recorded ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2834 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TMV reconstructed from micrographs recorded with the Falcon II DED | |||||||||
 Map data Map data | Cryo-EM structure of TMV reconstructed from micrographs recorded with the Falcon II DED | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tobacco mosaic virus / direct electron detectors / single particle helical reconstruction | |||||||||
| Biological species |   Tobacco mosaic virus Tobacco mosaic virus | |||||||||
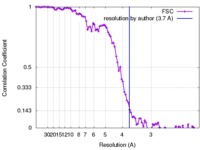
| Method | helical reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Fromm SA / Bharat TAM / Jakobi AJ / Hagen WJH / Sachse C | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2015 Journal: J Struct Biol / Year: 2015Title: Seeing tobacco mosaic virus through direct electron detectors. Authors: Simon A Fromm / Tanmay A M Bharat / Arjen J Jakobi / Wim J H Hagen / Carsten Sachse /   Abstract: With the introduction of direct electron detectors (DED) to the field of electron cryo-microscopy, a wave of atomic-resolution structures has become available. As the new detectors still require ...With the introduction of direct electron detectors (DED) to the field of electron cryo-microscopy, a wave of atomic-resolution structures has become available. As the new detectors still require comparative characterization, we have used tobacco mosaic virus (TMV) as a test specimen to study the quality of 3D image reconstructions from data recorded on the two direct electron detector cameras, K2 Summit and Falcon II. Using DED movie frames, we explored related image-processing aspects and compared the performance of micrograph-based and segment-based motion correction approaches. In addition, we investigated the effect of dose deposition on the atomic-resolution structure of TMV and show that radiation damage affects negative carboxyl chains first in a side-chain specific manner. Finally, using 450,000 asymmetric units and limiting the effects of radiation damage, we determined a high-resolution cryo-EM map at 3.35Å resolution. Here, we provide a comparative case study of highly ordered TMV recorded on different direct electron detectors to establish recording and processing conditions that enable structure determination up to 3.2Å in resolution using cryo-EM. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2834.map.gz emd_2834.map.gz | 23.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2834-v30.xml emd-2834-v30.xml emd-2834.xml emd-2834.xml | 10.6 KB 10.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2834_fsc.xml emd_2834_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_2834.png emd_2834.png | 175 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2834 http://ftp.pdbj.org/pub/emdb/structures/EMD-2834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2834 | HTTPS FTP |
-Validation report
| Summary document |  emd_2834_validation.pdf.gz emd_2834_validation.pdf.gz | 351.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2834_full_validation.pdf.gz emd_2834_full_validation.pdf.gz | 351 KB | Display | |
| Data in XML |  emd_2834_validation.xml.gz emd_2834_validation.xml.gz | 9.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2834 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2834 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2834 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2834 | HTTPS FTP |
-Related structure data
| Related structure data |  2833C  2835C  2836C  2837C  2838C  2839C  2840C  2841C  2842C  4udvC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10021 (Title: Tobacco Mosaic Virus Falcon II dataset including manually boxed helix coordinates EMPIAR-10021 (Title: Tobacco Mosaic Virus Falcon II dataset including manually boxed helix coordinatesData size: 28.0 Data #1: Tobacco Mosaic Virus micrographs [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2834.map.gz / Format: CCP4 / Size: 31.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2834.map.gz / Format: CCP4 / Size: 31.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of TMV reconstructed from micrographs recorded with the Falcon II DED | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
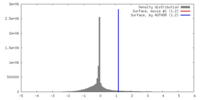
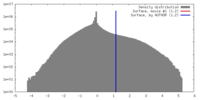
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tobacco Mosaic Virus
| Entire | Name:   Tobacco Mosaic Virus Tobacco Mosaic Virus |
|---|---|
| Components |
|
-Supramolecule #1000: Tobacco Mosaic Virus
| Supramolecule | Name: Tobacco Mosaic Virus / type: sample / ID: 1000 / Details: The sample was purified from infected leaves. / Oligomeric state: helical / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 39.5 MDa |
-Supramolecule #1: Tobacco mosaic virus
| Supramolecule | Name: Tobacco mosaic virus / type: virus / ID: 1 / Name.synonym: TMV / NCBI-ID: 12242 / Sci species name: Tobacco mosaic virus / Sci species strain: vulgare / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: TMV |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Theoretical: 39.5 MDa |
| Virus shell | Shell ID: 1 / Name: CP / Diameter: 180 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 5.5 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: 50 mM NaPO4 |
| Grid | Details: Quantifoil 2/2 Cu 300 mesh grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 95 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting at 283 kelvin Method: Blot for 2.5 seconds with force -15 ~10 seconds after sample application and with a drain time of 0.5 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: CTFTILT |
| Details | Nanoprobe mode, dose rate ~50 e-/px/s on the camera level |
| Date | Mar 14, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 28 / Average electron dose: 43 e/Å2 Details: each micrograph was recorded as a movie of 18 frames including the roll-in and roll-out frame which were discarded for image processing |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 131951 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)