[English] 日本語
 Yorodumi
Yorodumi- EMDB-28145: Cryo-EM structure of his-elemental paused elongation complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (out) | |||||||||
 Map data Map data | a postprocessed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase / cryo-EM structure / elemental pause / Escherichia coli / transcription / TRANSFERASE-DNA-RNA complex / transcriptional regulation / transcriptional pausing | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-directed RNA polymerase complex / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Kang JY / Chen J / Llewellyn E / Landick R / Darst SA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options. Authors: Jin Young Kang / Tatiana V Mishanina / Yu Bao / James Chen / Eliza Llewellyn / James Liu / Seth A Darst / Robert Landick /   Abstract: Transcriptional pausing underpins the regulation of cellular RNA synthesis, but its mechanism remains incompletely understood. Sequence-specific interactions of DNA and RNA with the dynamic, ...Transcriptional pausing underpins the regulation of cellular RNA synthesis, but its mechanism remains incompletely understood. Sequence-specific interactions of DNA and RNA with the dynamic, multidomain RNA polymerase (RNAP) trigger reversible conformational changes at pause sites that temporarily interrupt the nucleotide addition cycle. These interactions initially rearrange the elongation complex (EC) into an elemental paused EC (ePEC). ePECs can form longer-lived PECs by further rearrangements or interactions of diffusible regulators. For both bacterial and mammalian RNAPs, a half-translocated state in which the next DNA template base fails to load into the active site appears central to the ePEC. Some RNAPs also swivel interconnected modules that may stabilize the ePEC. However, it is unclear whether swiveling and half-translocation are requisite features of a single ePEC state or if multiple ePEC states exist. Here, we use cryo-electron microscopy (cryo-EM) analysis of ePECs with different RNA-DNA sequences combined with biochemical probes of ePEC structure to define an interconverting ensemble of ePEC states. ePECs occupy either pre- or half-translocated states but do not always swivel, indicating that difficulty in forming the posttranslocated state at certain RNA-DNA sequences may be the essence of the ePEC. The existence of multiple ePEC conformations has broad implications for transcriptional regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28145.map.gz emd_28145.map.gz | 60 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28145-v30.xml emd-28145-v30.xml emd-28145.xml emd-28145.xml | 33.6 KB 33.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28145.png emd_28145.png | 118 KB | ||
| Filedesc metadata |  emd-28145.cif.gz emd-28145.cif.gz | 9.2 KB | ||
| Others |  emd_28145_additional_1.map.gz emd_28145_additional_1.map.gz emd_28145_half_map_1.map.gz emd_28145_half_map_1.map.gz emd_28145_half_map_2.map.gz emd_28145_half_map_2.map.gz | 3.4 MB 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28145 http://ftp.pdbj.org/pub/emdb/structures/EMD-28145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28145 | HTTPS FTP |
-Related structure data
| Related structure data |  8ehaMC  8eg7C  8eg8C  8egbC  8eh8C  8eh9C  8ehfC  8ehiC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28145.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28145.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: a local-filtered map
| File | emd_28145_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a local-filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
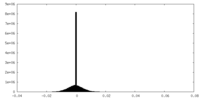
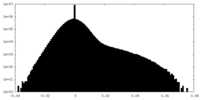
| Density Histograms |
-Half map: a first half map
| File | emd_28145_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a first half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: a second half map
| File | emd_28145_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a second half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : his-elemental paused elongation complex
+Supramolecule #1: his-elemental paused elongation complex
+Macromolecule #1: non-template DNA
+Macromolecule #2: template DNA
+Macromolecule #3: RNA
+Macromolecule #4: DNA-directed RNA polymerase subunit alpha
+Macromolecule #5: DNA-directed RNA polymerase subunit beta
+Macromolecule #6: DNA-directed RNA polymerase subunit beta'
+Macromolecule #7: DNA-directed RNA polymerase subunit omega
+Macromolecule #8: MAGNESIUM ION
+Macromolecule #9: CHAPSO
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: blot force 0, blot time 3.5 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 6939 / Average exposure time: 10.0 sec. / Average electron dose: 47.34 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||||||
| Output model |  PDB-8eha: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)