+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomograms of cryo-FIB milled WT dividing E. coli | ||||||||||||||||||
 Map data Map data | Cryo-electron tomogram of Cryo-FIB milled WT Dividing E. coli. Septation stage. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Cell division / CELL CYCLE | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | electron tomography / cryo EM | ||||||||||||||||||
 Authors Authors | Navarro PP / Vettiger A / Ananda VY / Montero Llopis P / Allolio C / Bernhardt TG / Chao LH | ||||||||||||||||||
| Funding support |  Switzerland, European Union, Switzerland, European Union,  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2022 Journal: Nat Microbiol / Year: 2022Title: Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli. Authors: Paula P Navarro / Andrea Vettiger / Virly Y Ananda / Paula Montero Llopis / Christoph Allolio / Thomas G Bernhardt / Luke H Chao /   Abstract: The bacterial division apparatus catalyses the synthesis and remodelling of septal peptidoglycan (sPG) to build the cell wall layer that fortifies the daughter cell poles. Understanding of this ...The bacterial division apparatus catalyses the synthesis and remodelling of septal peptidoglycan (sPG) to build the cell wall layer that fortifies the daughter cell poles. Understanding of this essential process has been limited by the lack of native three-dimensional views of developing septa. Here, we apply state-of-the-art cryogenic electron tomography (cryo-ET) and fluorescence microscopy to visualize the division site architecture and sPG biogenesis dynamics of the Gram-negative bacterium Escherichia coli. We identify a wedge-like sPG structure that fortifies the ingrowing septum. Experiments with strains defective in sPG biogenesis revealed that the septal architecture and mode of division can be modified to more closely resemble that of other Gram-negative (Caulobacter crescentus) or Gram-positive (Staphylococcus aureus) bacteria, suggesting that a conserved mechanism underlies the formation of different septal morphologies. Finally, analysis of mutants impaired in amidase activation (ΔenvC ΔnlpD) showed that cell wall remodelling affects the placement and stability of the cytokinetic ring. Taken together, our results support a model in which competition between the cell elongation and division machineries determines the shape of cell constrictions and the poles they form. They also highlight how the activity of the division system can be modulated to help generate the diverse array of shapes observed in the bacterial domain. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Cell wall synthesis and remodeling dynamics determine bacterial division site architecture and cell shape Authors: Navarro PP / Vettiger A / Ananda VY / Llopis PM / Allolio C / Bernhardt TG / Chao LH | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27479.map.gz emd_27479.map.gz | 141.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27479-v30.xml emd-27479-v30.xml emd-27479.xml emd-27479.xml | 34.4 KB 34.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27479.png emd_27479.png | 199.4 KB | ||
| Filedesc metadata |  emd-27479.cif.gz emd-27479.cif.gz | 4.6 KB | ||
| Others |  emd_27479_additional_1.map.gz emd_27479_additional_1.map.gz emd_27479_additional_10.map.gz emd_27479_additional_10.map.gz emd_27479_additional_11.map.gz emd_27479_additional_11.map.gz emd_27479_additional_2.map.gz emd_27479_additional_2.map.gz emd_27479_additional_3.map.gz emd_27479_additional_3.map.gz emd_27479_additional_4.map.gz emd_27479_additional_4.map.gz emd_27479_additional_5.map.gz emd_27479_additional_5.map.gz emd_27479_additional_6.map.gz emd_27479_additional_6.map.gz emd_27479_additional_7.map.gz emd_27479_additional_7.map.gz emd_27479_additional_8.map.gz emd_27479_additional_8.map.gz emd_27479_additional_9.map.gz emd_27479_additional_9.map.gz | 69.4 MB 224.5 MB 26.8 MB 266 MB 254 MB 59.1 MB 103.3 MB 20.7 MB 224.9 MB 24.5 MB 107.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27479 http://ftp.pdbj.org/pub/emdb/structures/EMD-27479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27479 | HTTPS FTP |
-Validation report
| Summary document |  emd_27479_validation.pdf.gz emd_27479_validation.pdf.gz | 593.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27479_full_validation.pdf.gz emd_27479_full_validation.pdf.gz | 593.3 KB | Display | |
| Data in XML |  emd_27479_validation.xml.gz emd_27479_validation.xml.gz | 4 KB | Display | |
| Data in CIF |  emd_27479_validation.cif.gz emd_27479_validation.cif.gz | 4.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27479 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27479 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27479 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27479 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27479.map.gz / Format: CCP4 / Size: 193 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_27479.map.gz / Format: CCP4 / Size: 193 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-electron tomogram of Cryo-FIB milled WT Dividing E. coli. Septation stage. | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 20.52 Å | ||||||||||||||||||||||||||||||||
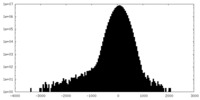
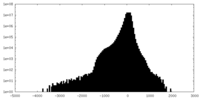
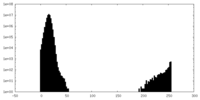
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E. coli. Pole.
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
+Additional map: Cryo-electron tomogram of Cryo-FIB milled WT Dividing E....
- Sample components
Sample components
-Entire : Dividing E. coli
| Entire | Name: Dividing E. coli |
|---|---|
| Components |
|
-Supramolecule #1: Dividing E. coli
| Supramolecule | Name: Dividing E. coli / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 9 |
|---|---|
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 294.15 K / Instrument: FEI VITROBOT MARK IV |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 5 / Focused ion beam - Current: 0.5 / Focused ion beam - Duration: 600 / Focused ion beam - Temperature: 88 K / Focused ion beam - Initial thickness: 1000 / Focused ion beam - Final thickness: 150 Focused ion beam - Details: The value given for _em_focused_ion_beam.instrument is Aquilos 2. This is not in a list of allowed values {'OTHER', 'DB235'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.5 µm / Nominal defocus min: 3.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 56 IMOD / Number images used: 56 |
|---|
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)