[English] 日本語
 Yorodumi
Yorodumi- EMDB-27201: EM map of the human UBR5 HECT-type E3 ubiquitin ligase in a dimer... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EM map of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form | |||||||||
 Map data Map data | Primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HECT / E3 ligase / Dimer / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationheterochromatin boundary formation / protein K29-linked ubiquitination / cytoplasm protein quality control by the ubiquitin-proteasome system / nuclear protein quality control by the ubiquitin-proteasome system / protein branched polyubiquitination / HECT-type E3 ubiquitin transferase / cytoplasm protein quality control / protein K11-linked ubiquitination / ubiquitin-ubiquitin ligase activity / DNA repair-dependent chromatin remodeling ...heterochromatin boundary formation / protein K29-linked ubiquitination / cytoplasm protein quality control by the ubiquitin-proteasome system / nuclear protein quality control by the ubiquitin-proteasome system / protein branched polyubiquitination / HECT-type E3 ubiquitin transferase / cytoplasm protein quality control / protein K11-linked ubiquitination / ubiquitin-ubiquitin ligase activity / DNA repair-dependent chromatin remodeling / progesterone receptor signaling pathway / protein K48-linked ubiquitination / ubiquitin binding / negative regulation of smoothened signaling pathway / positive regulation of protein import into nucleus / protein polyubiquitination / ubiquitin protein ligase activity / positive regulation of canonical Wnt signaling pathway / proteasome-mediated ubiquitin-dependent protein catabolic process / DNA repair / DNA damage response / positive regulation of gene expression / chromatin / perinuclear region of cytoplasm / protein-containing complex / RNA binding / zinc ion binding / nucleoplasm / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
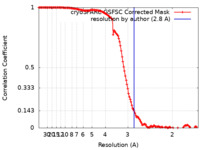
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Wang F / He Q / Lin G / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structure of the human UBR5 E3 ubiquitin ligase. Authors: Feng Wang / Qing He / Wenhu Zhan / Ziqi Yu / Efrat Finkin-Groner / Xiaojing Ma / Gang Lin / Huilin Li /  Abstract: The human UBR5 is a single polypeptide chain homology to E6AP C terminus (HECT)-type E3 ubiquitin ligase essential for embryonic development in mammals. Dysregulated UBR5 functions like an ...The human UBR5 is a single polypeptide chain homology to E6AP C terminus (HECT)-type E3 ubiquitin ligase essential for embryonic development in mammals. Dysregulated UBR5 functions like an oncoprotein to promote cancer growth and metastasis. Here, we report that UBR5 assembles into a dimer and a tetramer. Our cryoelectron microscopy (cryo-EM) structures reveal that two crescent-shaped UBR5 monomers assemble head to tail to form the dimer, and two dimers bind face to face to form the cage-like tetramer with all four catalytic HECT domains facing the central cavity. Importantly, the N-terminal region of one subunit and the HECT of the other form an "intermolecular jaw" in the dimer. We show the jaw-lining residues are important for function, suggesting that the intermolecular jaw functions to recruit ubiquitin-loaded E2 to UBR5. Further work is needed to understand how oligomerization regulates UBR5 ligase activity. This work provides a framework for structure-based anticancer drug development and contributes to a growing appreciation of E3 ligase diversity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27201.map.gz emd_27201.map.gz | 216.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27201-v30.xml emd-27201-v30.xml emd-27201.xml emd-27201.xml | 26.3 KB 26.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27201_fsc.xml emd_27201_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_27201.png emd_27201.png | 76.7 KB | ||
| Filedesc metadata |  emd-27201.cif.gz emd-27201.cif.gz | 8.6 KB | ||
| Others |  emd_27201_additional_1.map.gz emd_27201_additional_1.map.gz emd_27201_half_map_1.map.gz emd_27201_half_map_1.map.gz emd_27201_half_map_2.map.gz emd_27201_half_map_2.map.gz | 119.9 MB 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27201 http://ftp.pdbj.org/pub/emdb/structures/EMD-27201 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27201 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27201 | HTTPS FTP |
-Related structure data
| Related structure data |  8d4xMC  8e0qC  8ewiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27201.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27201.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: RAW map
| File | emd_27201_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RAW map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_27201_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_27201_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimer of human E3 ligase UBR5
| Entire | Name: Homodimer of human E3 ligase UBR5 |
|---|---|
| Components |
|
-Supramolecule #1: Homodimer of human E3 ligase UBR5
| Supramolecule | Name: Homodimer of human E3 ligase UBR5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: UBR5 expressed in insect cell |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 610 KDa |
-Macromolecule #1: E3 ubiquitin-protein ligase UBR5
| Macromolecule | Name: E3 ubiquitin-protein ligase UBR5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: HECT-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 310.604625 KDa |
| Recombinant expression | Organism: Insect expression vector pBlueBachsGCA1 (others) |
| Sequence | String: DYKDDDKMTS IHFVVHPLPG TEDQLNDRLR EVSEKLNKYN LNSHPPLNVL EQATIKQCVV GPNHAAFLLE DGRVCRIGFS VQPDRLELG KPDNNDGSKL NSNSGAGRTS RPGRTSDSPW FLSGSETLGR LAGNTLGSRW SSGVGGSGGG SSGRSSAGAR D SRRQTRVI ...String: DYKDDDKMTS IHFVVHPLPG TEDQLNDRLR EVSEKLNKYN LNSHPPLNVL EQATIKQCVV GPNHAAFLLE DGRVCRIGFS VQPDRLELG KPDNNDGSKL NSNSGAGRTS RPGRTSDSPW FLSGSETLGR LAGNTLGSRW SSGVGGSGGG SSGRSSAGAR D SRRQTRVI RTGRDRGSGL LGSQPQPVIP ASVIPEELIS QAQVVLQGKS RSVIIRELQR TNLDVNLAVN NLLSRDDEDG DD GDDTASE SYLPGEDLMS LLDADIHSAH PSVIIDADAM FSEDISYFGY PSFRRSSLSR LGSSRVLLLP LERDSELLRE RES VLRLRE RRWLDGASFD NERGSTSKEG EPNLDKKNTP VQSPVSLGED LQWWPDKDGT KFICIGALYS ELLAVSSKGE LYQW KWSES EPYRNAQNPS LHHPRATFLG LTNEKIVLLS ANSIRATVAT ENNKVATWVD ETLSSVASKL EHTAQTYSEL QGERI VSLH CCALYTCAQL ENSLYWWGVV PFSQRKKMLE KARAKNKKPK SSAGISSMPN ITVGTQVCLR NNPLYHAGAV AFSISA GIP KVGVLMESVW NMNDSCRFQL RSPESLKNME KASKTTEAKP ESKQEPVKTE MGPPPSPAST CSDASSIASS ASMPYKR RR STPAPKEEEK VNEEQWSLRE VVFVEDVKNV PVGKVLKVDG AYVAVKFPGT SSNTNCQNSS GPDADPSSLL QDCRLLRI D ELQVVKTGGT PKVPDCFQRT PKKLCIPEKT EILAVNVDSK GVHAVLKTGN WVRYCIFDLA TGKAEQENNF PTSSIAFLG QNERNVAIFT AGQESPIILR DGNGTIYPMA KDCMGGIRDP DWLDLPPISS LGMGVHSLIN LPANSTIKKK AAVIIMAVEK QTLMQHILR CDYEACRQYL MNLEQAVVLE QNLQMLQTFI SHRCDGNRNI LHACVSVCFP TSNKETKEEE EAERSERNTF A ERLSAVEA IANAISVVSS NGPGNRAGSS SSRSLRLREM MRRSLRAAGL GRHEAGASSS DHQDPVSPPI APPSWVPDPP AM DPDGDID FILAPAVGSL TTAATGTGQG PSTSTIPGPS TEPSVVESKD RKANAHFILK LLCDSVVLQP YLRELLSAKD ARG MTPFMS AVSGRAYPAA ITILETAQKI AKAEISSSEK EEDVFMGMVC PSGTNPDDSP LYVLCCNDTC SFTWTGAEHI NQDI FECRT CGLLESLCCC TECARVCHKG HDCKLKRTSP TAYCDCWEKC KCKTLIAGQK SARLDLLYRL LTATNLVTLP NSRGE HLLL FLVQTVARQT VEHCQYRPPR IREDRNRKTA SPEDSDMPDH DLEPPRFAQL ALERVLQDWN ALKSMIMFGS QENKDP LSA SSRIGHLLPE EQVYLNQQSG TIRLDCFTHC LIVKCTADIL LLDTLLGTLV KELQNKYTPG RREEAIAVTM RFLRSVA RV FVILSVEMAS SKKKNNFIPQ PIGKCKRVFQ ALLPYAVEEL CNVAESLIVP VRMGIARPTA PFTLASTSID AMQGSEEL F SVEPLPPRPS SDQSSSSSQS QSSYIIRNPQ QRRISQSQPV RGRDEEQDDI VSADVEEVEV VEGVAGEEDH HDEQEEHGE ENAEAEGQHD EHDEDGSDME LDLLAAAETE SDSESNHSNQ DNASGRRSVV TAATAGSEAG ASSVPAFFSE DDSQSNDSSD SDSSSSQSD DIEQETFMLD EPLERTTNSS HANGAAQAPR SMQWAVRNTQ HQRAASTAPS STSTPAASSA GLIYIDPSNL R RSGTISTS AAAAAAALEA SNASSYLTSA SSLARAYSIV IRQISDLMGL IPKYNHLVYS QIPAAVKLTY QDAVNLQNYV EE KLIPTWN WMVSIMDSTE AQLRYGSALA SAGDPGHPNH PLHASQNSAR RERMTAREEA SLRTLEGRRR ATLLSARQGM MSA RGDFLN YALSLMRSHN DEHSDVLPVL DVCSLKHVAY VFQALIYWIK AMNQQTTLDT PQLERKRTRE LLELGIDNED SEHE NDDDT NQSATLNDKD DDSLPAETGQ NHPFFRRSDS MTFLGCIPPN PFEVPLAEAI PLADQPHLLQ PNARKEDLFG RPSQG LYSS SASSGKCLME VTVDRNCLEV LPTKMSYAAN LKNVMNMQNR QKKEGEEQPV LPEETESSKP GPSAHDLAAQ LKSSLL AEI GLTESEGPPL TSFRPQCSFM GMVISHDMLL GRWRLSLELF GRVFMEDVGA EPGSILTELG GFEVKESKFR REMEKLR NQ QSRDLSLEVD RDRDLLIQQT MRQLNNHFGR RCATTPMAVH RVKVTFKDEP GEGSGVARSF YTAIAQAFLS NEKLPNLE C IQNANKGTHT SLMQRLRNRG ERDRERERER EMRRSSGLRA GSRRDRDRDF RRQLSIDTRP FRPASEGNPS DDPEPLPAH RQALGERLYP RVQAMQPAFA SKITGMLLEL SPAQLLLLLA SEDSLRARVD EAMELIIAHG RENGADSILD LGLVDSSEKV QQENRKRHG SSRSVVDMDL DDTDDGDDNA PLFYQPGKRG FYTPRPGKNT EARLNCFRNI GRILGLCLLQ NELCPITLNR H VIKVLLGR KVNWHDFAFF DPVMYESLRQ LILASQSSDA DAVFSAMDLA FAIDLCKEEG GGQVELIPNG VNIPVTPQNV YE YVRKYAE HRMLVVAEQP LHAMRKGLLD VLPKNSLEDL TAEDFRLLVN GCGEVNVQML ISFTSFNDES GENAEKLLQF KRW FWSIVE KMSMTERQDL VYFWTSSPSL PASEEGFQPM PSITIRPPDD QHLPTANTCI SRLYVPLYSS KQILKQKLLL AIKT KNFGF V UniProtKB: E3 ubiquitin-protein ligase UBR5 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: GOLD / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV / Details: blot 2S, blot forth 2. | ||||||||||||
| Details | freshly purified UBR5 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 193.0 K / Max: 193.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.0 e/Å2 Details: Images were collected in movie-mode at 75 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 90.94 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-8d4x: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)