[English] 日本語
 Yorodumi
Yorodumi- EMDB-26780: Focused map of the closed state GEF domain of Gea2 closed/open co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused map of the closed state GEF domain of Gea2 closed/open conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Muccini A / Fromme JC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Structural basis for activation of Arf1 at the Golgi complex. Authors: Arnold J Muccini / Margaret A Gustafson / J Christopher Fromme /  Abstract: The Golgi complex is the central sorting station of the eukaryotic secretory pathway. Traffic through the Golgi requires activation of Arf guanosine triphosphatases that orchestrate cargo sorting and ...The Golgi complex is the central sorting station of the eukaryotic secretory pathway. Traffic through the Golgi requires activation of Arf guanosine triphosphatases that orchestrate cargo sorting and vesicle formation by recruiting an array of effector proteins. Arf activation and Golgi membrane association is controlled by large guanine nucleotide exchange factors (GEFs) possessing multiple conserved regulatory domains. Here we present cryoelectron microscopy (cryoEM) structures of full-length Gea2, the yeast paralog of the human Arf-GEF GBF1, that reveal the organization of these regulatory domains and explain how Gea2 binds to the Golgi membrane surface. We find that the GEF domain adopts two different conformations compatible with different stages of the Arf activation reaction. The structure of a Gea2-Arf1 activation intermediate suggests that the movement of the GEF domain primes Arf1 for membrane insertion upon guanosine triphosphate binding. We propose that conformational switching of Gea2 during the nucleotide exchange reaction promotes membrane insertion of Arf1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26780.map.gz emd_26780.map.gz | 431.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26780-v30.xml emd-26780-v30.xml emd-26780.xml emd-26780.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26780.png emd_26780.png | 20.8 KB | ||
| Masks |  emd_26780_msk_1.map emd_26780_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Others |  emd_26780_half_map_1.map.gz emd_26780_half_map_1.map.gz emd_26780_half_map_2.map.gz emd_26780_half_map_2.map.gz | 40.8 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26780 http://ftp.pdbj.org/pub/emdb/structures/EMD-26780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26780 | HTTPS FTP |
-Validation report
| Summary document |  emd_26780_validation.pdf.gz emd_26780_validation.pdf.gz | 557.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26780_full_validation.pdf.gz emd_26780_full_validation.pdf.gz | 556.7 KB | Display | |
| Data in XML |  emd_26780_validation.xml.gz emd_26780_validation.xml.gz | 11.4 KB | Display | |
| Data in CIF |  emd_26780_validation.cif.gz emd_26780_validation.cif.gz | 13.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26780 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26780 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26780 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26780 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26780.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26780.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.664 Å | ||||||||||||||||||||||||||||||||||||
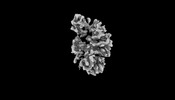
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26780_msk_1.map emd_26780_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
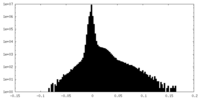
| Density Histograms |
-Half map: #1
| File | emd_26780_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26780_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Gea2
| Entire | Name: Gea2 |
|---|---|
| Components |
|
-Supramolecule #1: Gea2
| Supramolecule | Name: Gea2 / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 101014 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)