[English] 日本語
 Yorodumi
Yorodumi- EMDB-26347: Human CST bound to full-length DNA polymerase alpha-primase in a ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human CST bound to full-length DNA polymerase alpha-primase in a recruitment state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
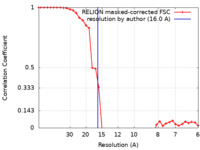
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Cai SW / Zinder JC / Svetlov V / Bush MW / Nudler E / Walz T / de Lange T | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Cryo-EM structure of the human CST-Polα/primase complex in a recruitment state. Authors: Sarah W Cai / John C Zinder / Vladimir Svetlov / Martin W Bush / Evgeny Nudler / Thomas Walz / Titia de Lange /  Abstract: The CST-Polα/primase complex is essential for telomere maintenance and functions to counteract resection at double-strand breaks. We report a 4.6-Å resolution cryo-EM structure of human CST- ...The CST-Polα/primase complex is essential for telomere maintenance and functions to counteract resection at double-strand breaks. We report a 4.6-Å resolution cryo-EM structure of human CST-Polα/primase, captured prior to catalysis in a recruitment state stabilized by chemical cross-linking. Our structure reveals an evolutionarily conserved interaction between the C-terminal domain of the catalytic POLA1 subunit and an N-terminal expansion in metazoan CTC1. Cross-linking mass spectrometry and negative-stain EM analysis provide insight into CST binding by the flexible POLA1 N-terminus. Finally, Coats plus syndrome disease mutations previously characterized to disrupt formation of the CST-Polα/primase complex map to protein-protein interfaces observed in the recruitment state. Together, our results shed light on the architecture and stoichiometry of the metazoan fill-in machinery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26347.map.gz emd_26347.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26347-v30.xml emd-26347-v30.xml emd-26347.xml emd-26347.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26347_fsc.xml emd_26347_fsc.xml | 3.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_26347.png emd_26347.png | 49.5 KB | ||
| Others |  emd_26347_half_map_1.map.gz emd_26347_half_map_1.map.gz emd_26347_half_map_2.map.gz emd_26347_half_map_2.map.gz | 2.8 MB 2.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26347 http://ftp.pdbj.org/pub/emdb/structures/EMD-26347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26347 | HTTPS FTP |
-Related structure data
| Related structure data |  7u5cC C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-11130 (Title: Single particle cryo-EM of the human CST•Polα/Primase (POLA1 FL) complex in a recruitment state EMPIAR-11130 (Title: Single particle cryo-EM of the human CST•Polα/Primase (POLA1 FL) complex in a recruitment stateData size: 317.1 Data #1: Motion corrected micrographs collected on Talos Arctica of CST-Pola/Primase FL complex in recruitment state [micrographs - single frame] Data #2: Motion corrected micrographs collected on Titan Krios of CST-Pola/Primase FL complex in recruitment state [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26347.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26347.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3 Å | ||||||||||||||||||||||||||||||||||||
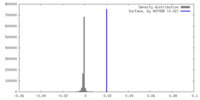
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_26347_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
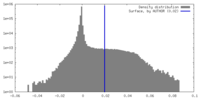
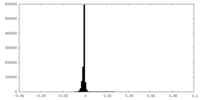
| Density Histograms |
-Half map: #2
| File | emd_26347_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
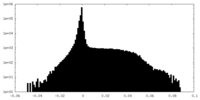
| Density Histograms |
- Sample components
Sample components
-Entire : CST bound to full-length DNA polymerase alpha-primase and ssDNA
| Entire | Name: CST bound to full-length DNA polymerase alpha-primase and ssDNA |
|---|---|
| Components |
|
-Supramolecule #1: CST bound to full-length DNA polymerase alpha-primase and ssDNA
| Supramolecule | Name: CST bound to full-length DNA polymerase alpha-primase and ssDNA type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all / Details: GraFix cross-linked |
|---|---|
| Molecular weight | Theoretical: 540 KDa |
-Macromolecule #1: DNA primase catalytic subunit
| Macromolecule | Name: DNA primase catalytic subunit / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: METFDPTELP ELLKLYYRRL FPYSQYYRWL NYGGVIKNYF QHREFSFTLK DDIYIRYQSF NNQSDLEKEM QKMNPYKIDI GAVYSHRPNQ HNTVKLGAFQ AQEKELVFDI DMTDYDDVRR CCSSADICPK CWTLMTMAIR IIDRALKEDF GFKHRLWVYS GRRGVHCWVC ...String: METFDPTELP ELLKLYYRRL FPYSQYYRWL NYGGVIKNYF QHREFSFTLK DDIYIRYQSF NNQSDLEKEM QKMNPYKIDI GAVYSHRPNQ HNTVKLGAFQ AQEKELVFDI DMTDYDDVRR CCSSADICPK CWTLMTMAIR IIDRALKEDF GFKHRLWVYS GRRGVHCWVC DESVRKLSSA VRSGIVEYLS LVKGGQDVKK KVHLSEKIHP FIRKSINIIK KYFEEYALVN QDILENKESW DKILALVPET IHDELQQSFQ KSHNSLQRWE HLKKVASRYQ NNIKNDKYGP WLEWEIMLQY CFPRLDINVS KGINHLLKSP FSVHPKTGRI SVPIDLQKVD QFDPFTVPTI SFICRELDAI STNEEEKEEN EAESDVKHRT RDYKKTSLAP YVKVFEHFLE NLDKSRKGEL LKKSDLQKDF |
-Macromolecule #2: DNA primase large subunit PRIM2
| Macromolecule | Name: DNA primase large subunit PRIM2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MEFSGRKWRK LRLAGDQRNA SYPHCLQFYL QPPSENISLI EFENLAIDRV KLLKSVENLG VSYVKGTEQY QSKLESELRK LKFSYRENLE DEYEPRRRDH ISHFILRLAY CQSEELRRWF IQQEMDLLRF RFSILPKDKI QDFLKDSQLQ FEAISDEEKT LREQEIVASS ...String: MEFSGRKWRK LRLAGDQRNA SYPHCLQFYL QPPSENISLI EFENLAIDRV KLLKSVENLG VSYVKGTEQY QSKLESELRK LKFSYRENLE DEYEPRRRDH ISHFILRLAY CQSEELRRWF IQQEMDLLRF RFSILPKDKI QDFLKDSQLQ FEAISDEEKT LREQEIVASS PSLSGLKLGF ESIYKIPFAD ALDLFRGRKV YLEDGFAYVP LKDIVAIILN EFRAKLSKAL ALTARSLPAV QSDERLQPLL NHLSHSYTGQ DYSTQGNVGK ISLDQIDLLS TKSFPPCMRQ LHKALRENHH LRHGGRMQYG LFLKGIGLTL EQALQFWKQE FIKGKMDPDK FDKGYSYNIR HSFGKEGKRT DYTPFSCLKI ILSNPPSQGD YHGCPFRHSD PELLKQKLQS YKISPGGISQ ILDLVKGTHY QVACQKYFEM IHNVDDCGFS LNHPNQFFCE SQRILNGGKD IKKEPIQPET PQPKPSVQKT KDASSALASL NSSLEMDMEG LEDYFSEDS |
-Macromolecule #3: DNA polymerase alpha catalytic subunit POLA1
| Macromolecule | Name: DNA polymerase alpha catalytic subunit POLA1 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAPVHGDDSL SDSGSFVSSR ARREKKSKKG RQEALERLKK AKAGEKYKYE VEDFTGVYEE VDEEQYSKLV QARQDDDWIV DDDGIGYVED GREIFDDDLE DDALDADEKG KDGKARNKDK RNVKKLAVTK PNNIKSMFIA CAGKKTADKA VDLSKDGLLG DILQDLNTET ...String: MAPVHGDDSL SDSGSFVSSR ARREKKSKKG RQEALERLKK AKAGEKYKYE VEDFTGVYEE VDEEQYSKLV QARQDDDWIV DDDGIGYVED GREIFDDDLE DDALDADEKG KDGKARNKDK RNVKKLAVTK PNNIKSMFIA CAGKKTADKA VDLSKDGLLG DILQDLNTET PQITPPPVMI LKKKRSIGAS PNPFSVHTAT AVPSGKIASP VSRKEPPLTP VPLKRAEFAG DDVQVESTEE EQESGAMEFE DGDFDEPMEV EEVDLEPMAA KAWDKESEPA EEVKQEADSG KGTVSYLGSF LPDVSCWDID QEGDSSFSVQ EVQVDSSHLP LVKGADEEQV FHFYWLDAYE DQYNQPGVVF LFGKVWIESA ETHVSCCVMV KNIERTLYFL PREMKIDLNT GKETGTPISM KDVYEEFDEK IATKYKIMKF KSKPVEKNYA FEIPDVPEKS EYLEVKYSAE MPQLPQDLKG ETFSHVFGTN TSSLELFLMN RKIKGPCWLE VKSPQLLNQP VSWCKVEAMA LKPDLVNVIK DVSPPPLVVM AFSMKTMQNA KNHQNEIIAM AALVHHSFAL DKAAPKPPFQ SHFCVVSKPK DCIFPYAFKE VIEKKNVKVE VAATERTLLG FFLAKVHKID PDIIVGHNIY GFELEVLLQR INVCKAPHWS KIGRLKRSNM PKLGGRSGFG ERNATCGRMI CDVEISAKEL IRCKSYHLSE LVQQILKTER VVIPMENIQN MYSESSQLLY LLEHTWKDAK FILQIMCELN VLPLALQITN IAGNIMSRTL MGGRSERNEF LLLHAFYENN YIVPDKQIFR KPQQKLGDED EEIDGDTNKY KKGRKKAAYA GGLVLDPKVG FYDKFILLLD FNSLYPSIIQ EFNICFTTVQ RVASEAQKVT EDGEQEQIPE LPDPSLEMGI LPREIRKLVE RRKQVKQLMK QQDLNPDLIL QYDIRQKALK LTANSMYGCL GFSYSRFYAK PLAALVTYKG REILMHTKEM VQKMNLEVIY GDTDSIMINT NSTNLEEVFK LGNKVKSEVN KLYKLLEIDI DGVFKSLLLL KKKKYAALVV EPTSDGNYVT KQELKGLDIV RRDWCDLAKD TGNFVIGQIL SDQSRDTIVE NIQKRLIEIG ENVLNGSVPV SQFEINKALT KDPQDYPDKK SLPHVHVALW INSQGGRKVK AGDTVSYVIC QDGSNLTASQ RAYAPEQLQK QDNLTIDTQY YLAQQIHPVV ARICEPIDGI DAVLIATWLG LDPTQFRVHH YHKDEENDAL LGGPAQLTDE EKYRDCERFK CPCPTCGTEN IYDNVFDGSG TDMEPSLYRC SNIDCKASPL TFTVQLSNKL IMDIRRFIKK YYDGWLICEE PTCRNRTRHL PLQFSRTGPL CPACMKATLQ PEYSDKSLYT QLCFYRYIFD AECALEKLTT DHEKDKLKKQ FFTPKVLQDY RKLKNTAEQF LSRSGYSEVN LSKLFAGCAV KS |
-Macromolecule #4: DNA polymerase alpha B subunit POLA2
| Macromolecule | Name: DNA polymerase alpha B subunit POLA2 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSASAQQLAE ELQIFGLDCE EALIEKLVEL CVQYGQNEEG MVGELIAFCT STHKVGLTSE ILNSFEHEFL SKRLSKARHS TCKDSGHAGA RDIVSIQELI EVEEEEEILL NSYTTPSKGS QKRAISTPET PLTKRSVSTR SPHQLLSPSS FSPSATPSQK YNSRSNRGEV ...String: MSASAQQLAE ELQIFGLDCE EALIEKLVEL CVQYGQNEEG MVGELIAFCT STHKVGLTSE ILNSFEHEFL SKRLSKARHS TCKDSGHAGA RDIVSIQELI EVEEEEEILL NSYTTPSKGS QKRAISTPET PLTKRSVSTR SPHQLLSPSS FSPSATPSQK YNSRSNRGEV VTSFGLAQGV SWSGRGGAGN ISLKVLGCPE ALTGSYKSMF QKLPDIREVL TCKIEELGSE LKEHYKIEAF TPLLAPAQEP VTLLGQIGCD SNGKLNNKSV ILEGDREHSS GAQIPVDLSE LKEYSLFPGQ VVIMEGINTT GRKLVATKLY EGVPLPFYQP TEEDADFEQS MVLVACGPYT TSDSITYDPL LDLIAVINHD RPDVCILFGP FLDAKHEQVE NCLLTSPFED IFKQCLRTII EGTRSSGSHL VFVPSLRDVH HEPVYPQPPF SYSDLSREDK KQVQFVSEPC SLSINGVIFG LTSTDLLFHL GAEEISSSSG TSDRFSRILK HILTQRSYYP LYPPQEDMAI DYESFYVYAQ LPVTPDVLII PSELRYFVKD VLGCVCVNPG RLTKGQVGGT FARLYLRRPA ADGAERQSPC IAVQVVRI |
-Macromolecule #5: CST complex subunit Ctc1
| Macromolecule | Name: CST complex subunit Ctc1 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAGRAQVPS SEQAWLEDAQ VFIQKTLCPA VKEPNVQLTP LVIDCVKTVW LSQGRNQGST LPLSYSFVSV QDLKTHQRLP CCSHLSWSSS AYQAWAQEAG PNGNPLPREQ LLLLGTLTDL SADLEQECRN GSLYVRDNTG VLSCELIDLD LSWLGHLFLF PRWSYLPPAR ...String: MAAGRAQVPS SEQAWLEDAQ VFIQKTLCPA VKEPNVQLTP LVIDCVKTVW LSQGRNQGST LPLSYSFVSV QDLKTHQRLP CCSHLSWSSS AYQAWAQEAG PNGNPLPREQ LLLLGTLTDL SADLEQECRN GSLYVRDNTG VLSCELIDLD LSWLGHLFLF PRWSYLPPAR WNSSGEGHLE LWDAPVPVFP LTISPGPVTP IPVLYPESAS CLLRLRNKLR GVQRNLAGSL VRLSALVKSK QKAYFILSLG RSHPAVTHVS IIVQVPAQLV WHRALRPGTA YVLTELRVSK IRGQRQHVWM TSQSSRLLLL KPECVQELEL ELEGPLLEAD PKPLPMPSNS EDKKDPESLV RYSRLLSYSG AVTGVLNEPA GLYELDGQLG LCLAYQQFRG LRRVMRPGVC LQLQDVHLLQ SVGGGTRRPV LAPCLRGAVL LQSFSRQKPG AHSSRQAYGA SLYEQLVWER QLGLPLYLWA TKALEELACK LCPHVLRHHQ FLQHSSPGSP SLGLQLLAPT LDLLAPPGSP VRNAHNEILE EPHHCPLQKY TRLQTPSSFP TLATLKEEGQ RKAWASFDPK ALLPLPEASY LPSCQLNRRL AWSWLCLLPS AFCPAQVLLG VLVASSHKGC LQLRDQSGSL PCLLLAKHSQ PLSDPRLIGC LVRAERFQLI VERDVRSSFP SWKELSMPGF IQKQQARVYV QFFLADALIL PVPRPCLHSA TPSTPQTDPT GPEGPHLGQS RLFLLCHKEA LMKRNFCVPP GASPEVPKPA LSFYVLGSWL GGTQRKEGTG WGLPEPQGND DNDQKVHLIF FGSSVRWFEF LHPGQVYRLI APGPATPMLF EKDGSSCISR RPLELAGCAS CLTVQDNWTL ELESSQDIQD VLDANKSLPE SSLTDLLSDN FTDSLVSFSA EILSRTLCEP LVASLWMKLG NTGAMRRCVK LTVALETAEC EFPPHLDVYI EDPHLPPSLG LLPGARVHFS QLEKRVSRSH NVYCCFRSST YVQVLSFPPE TTISIPLPHI YLAELLQGGQ SPFQATASCH IVSVFSLQLF WVCAYCTSIC RQGKCTRLGS TCPTQTAISQ AIIRLLVEDG TAEAVVTCRN HHVAAALGLC PREWASLLDF VQVPGRVVLQ FAGPGAQLES SARVDEPMTM FLWTLCTSPS VLRPIVLSFE LERKPSKIVP LEPPRLQRFQ CGELPFLTHV NPRLRLSCLS IRESEYSSSL GILASSC |
-Macromolecule #6: CST complex subunit Stn1
| Macromolecule | Name: CST complex subunit Stn1 / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MQPGSSRCEE ETPSLLWGLD PVFLAFAKLY IRDILDMKES RQVPGVFLYN GHPIKQVDVL GTVIGVRERD AFYSYGVDDS TGVINCICWK KLNTESVSAA PSAARELSLT SQLKKLQETI EQKTKIEIGD TIRVRGSIRT YREEREIHAT TYYKVDDPVW NIQIARMLEL ...String: MQPGSSRCEE ETPSLLWGLD PVFLAFAKLY IRDILDMKES RQVPGVFLYN GHPIKQVDVL GTVIGVRERD AFYSYGVDDS TGVINCICWK KLNTESVSAA PSAARELSLT SQLKKLQETI EQKTKIEIGD TIRVRGSIRT YREEREIHAT TYYKVDDPVW NIQIARMLEL PTIYRKVYDQ PFHSSALEKE EALSNPGALD LPSLTSLLSE KAKEFLMENR VQSFYQQELE MVESLLSLAN QPVIHSASSD QVNFKKDTTS KAIHSIFKNA IQLLQEKGLV FQKDDGFDNL YYVTREDKDL HRKIHRIIQQ DCQKPNHMEK GCHFLHILAC ARLSIRPGLS EAVLQQVLEL LEDQSDIVST MEHYYTAF |
-Macromolecule #7: CST complex subunit Ten1
| Macromolecule | Name: CST complex subunit Ten1 / type: protein_or_peptide / ID: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MMLPKPGTYY LPWEVSAGQV PDGSTLRTFG RLCLYDMIQS RVTLMAQHGS DQHQVLVCTK LVEPFHAQVG SLYIVLGELQ HQQDRGSVVK ARVLTCVEGM NLPLLEQAIR EQRLYKQERG GSQ |
-Macromolecule #8: Telomeric G-strand ssDNA (3 repeats)
| Macromolecule | Name: Telomeric G-strand ssDNA (3 repeats) / type: dna / ID: 8 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TALOS ARCTICA |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)