+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C2-symmetric single-particle cryo-EM map of T. vaginalis FDPF3 | |||||||||
 Map data Map data | C2-symmetric, sharpened, masked map of TvFDPF3 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | oxygen scavenging / fmn / nadh / flavodiiron protein / OXIDOREDUCTASE | |||||||||
| Biological species |  Trichomonas vaginalis (eukaryote) Trichomonas vaginalis (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.8 Å | |||||||||
 Authors Authors | Bell TA / Chao LH | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: A natural fusion of flavodiiron, rubredoxin, and rubredoxin oxidoreductase domains is a self-sufficient water-forming oxidase of Trichomonas vaginalis. Authors: Evana N Abdulaziz / Tristan A Bell / Bazlur Rashid / Mina L Heacock / Tarik Begic / Owen S Skinner / Mohammad A Yaseen / Luke H Chao / Vamsi K Mootha / Antonio J Pierik / Valentin Cracan /   Abstract: Microaerophilic pathogens such as Giardia lamblia, Entamoeba histolytica, and Trichomonas vaginalis have robust oxygen consumption systems to detoxify oxygen and maintain intracellular redox balance. ...Microaerophilic pathogens such as Giardia lamblia, Entamoeba histolytica, and Trichomonas vaginalis have robust oxygen consumption systems to detoxify oxygen and maintain intracellular redox balance. This oxygen consumption results from HO-forming NADH oxidase (NOX) activity of two distinct flavin-containing systems: HO-forming NOXes and multicomponent flavodiiron proteins (FDPs). Neither system is membrane bound, and both recycle NADH into oxidized NAD while simultaneously removing O from the local environment. However, little is known about the specific contributions of these systems in T. vaginalis. In this study, we use bioinformatics and biochemical analyses to show that T. vaginalis lacks a NOX-like enzyme and instead harbors three paralogous genes (FDPF1-3), each encoding a natural fusion product between the N-terminal FDP, central rubredoxin (Rb), and C-terminal NADH:Rb oxidoreductase domains. Unlike a "stand-alone" FDP that lacks Rb and oxidoreductase domains, this natural fusion protein with fully populated flavin redox centers directly accepts reducing equivalents of NADH to catalyze the four-electron reduction of oxygen to water within a single polypeptide with an extremely high turnover. Furthermore, using single-particle cryo-EM, we present structural insights into the spatial organization of the FDP core within this multidomain fusion protein. Together, these results contribute to our understanding of systems that allow protozoan parasites to maintain optimal redox balance and survive transient exposure to oxic conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25787.map.gz emd_25787.map.gz | 3.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25787-v30.xml emd-25787-v30.xml emd-25787.xml emd-25787.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25787_fsc.xml emd_25787_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25787.png emd_25787.png | 83.3 KB | ||
| Filedesc metadata |  emd-25787.cif.gz emd-25787.cif.gz | 5 KB | ||
| Others |  emd_25787_additional_1.map.gz emd_25787_additional_1.map.gz emd_25787_half_map_1.map.gz emd_25787_half_map_1.map.gz emd_25787_half_map_2.map.gz emd_25787_half_map_2.map.gz | 59.6 MB 48.5 MB 48.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25787 http://ftp.pdbj.org/pub/emdb/structures/EMD-25787 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25787 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25787 | HTTPS FTP |
-Validation report
| Summary document |  emd_25787_validation.pdf.gz emd_25787_validation.pdf.gz | 684.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25787_full_validation.pdf.gz emd_25787_full_validation.pdf.gz | 684.4 KB | Display | |
| Data in XML |  emd_25787_validation.xml.gz emd_25787_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_25787_validation.cif.gz emd_25787_validation.cif.gz | 21.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25787 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25787 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25787 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25787 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10895 (Title: C2-symmetric single-particle cryo-EM map of T. vaginalis FDPF3 - unaligned multi-frame micrographs EMPIAR-10895 (Title: C2-symmetric single-particle cryo-EM map of T. vaginalis FDPF3 - unaligned multi-frame micrographsData size: 1.4 TB Data #1: Unaligned multi-frame micrographs of recombinantly expressed and purified T. vaginalis FDPF3 [picked particles - multiframe - unprocessed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25787.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25787.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2-symmetric, sharpened, masked map of TvFDPF3 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
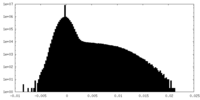
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: C2-symmetric, sharpened, unmasked map of TvFDPF3
| File | emd_25787_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2-symmetric, sharpened, unmasked map of TvFDPF3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
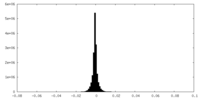
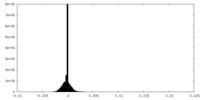
| Density Histograms |
-Half map: Half-map 1 from masked refinement of C2-symmetric map...
| File | emd_25787_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 from masked refinement of C2-symmetric map prior to sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
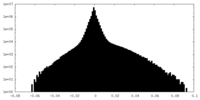
| Density Histograms |
-Half map: Half-map 2 from masked refinement of C2-symmetric map...
| File | emd_25787_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 from masked refinement of C2-symmetric map prior to sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
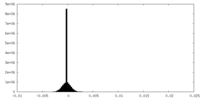
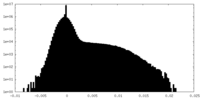
| Density Histograms |
- Sample components
Sample components
-Entire : Dimeric assembly of TvFDPF3 N-terminal FDP-like domains
| Entire | Name: Dimeric assembly of TvFDPF3 N-terminal FDP-like domains |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric assembly of TvFDPF3 N-terminal FDP-like domains
| Supramolecule | Name: Dimeric assembly of TvFDPF3 N-terminal FDP-like domains type: complex / ID: 1 / Parent: 0 / Details: Resolved portion of full-lengh TvFDPF3 |
|---|---|
| Source (natural) | Organism:  Trichomonas vaginalis (eukaryote) / Strain: C-1:NIH Trichomonas vaginalis (eukaryote) / Strain: C-1:NIH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 3.9 kPa Details: Grid was washed twice with chloroform and air dried prior to glow discharge and sample application. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting condition: 5 s at +15 blotting force.. | |||||||||
| Details | Monodisperse sample |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 7398 / Average electron dose: 54.5 e/Å2 / Details: Images were collected as 51-frame movies. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)