+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA-induced tau amyloid fibril | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | amyloid fibril / complex / PROTEIN FIBRIL-RNA complex | |||||||||
| Function / homology | Activation of AMPK downstream of NMDARs / PKR-mediated signaling / Isoform Tau-F of Microtubule-associated protein tau Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
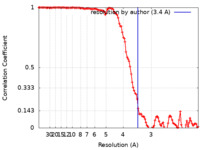
| Method | helical reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Abskharon R / Sawaya MR | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Cryo-EM structure of RNA-induced tau fibrils reveals a small C-terminal core that may nucleate fibril formation. Authors: Romany Abskharon / Michael R Sawaya / David R Boyer / Qin Cao / Binh A Nguyen / Duilio Cascio / David S Eisenberg /  Abstract: In neurodegenerative diseases including Alzheimer’s and amyotrophic lateral sclerosis, proteins that bind RNA are found in aggregated forms in autopsied brains. Evidence suggests that RNA aids ...In neurodegenerative diseases including Alzheimer’s and amyotrophic lateral sclerosis, proteins that bind RNA are found in aggregated forms in autopsied brains. Evidence suggests that RNA aids nucleation of these pathological aggregates; however, the mechanism has not been investigated at the level of atomic structure. Here, we present the 3.4-Å resolution structure of fibrils of full-length recombinant tau protein in the presence of RNA, determined by electron cryomicroscopy (cryo-EM). The structure reveals the familiar in-register cross-β amyloid scaffold but with a small fibril core spanning residues Glu391 to Ala426, a region disordered in the fuzzy coat in all previously studied tau polymorphs. RNA is bound on the fibril surface to the positively charged residues Arg406 and His407 and runs parallel to the fibril axis. The fibrils dissolve when RNase is added, showing that RNA is necessary for fibril integrity. While this structure cannot exist simultaneously with the tau fibril structures extracted from patients’ brains, it could conceivably account for the nucleating effects of RNA cofactors followed by remodeling as fibrils mature. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25364.map.gz emd_25364.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25364-v30.xml emd-25364-v30.xml emd-25364.xml emd-25364.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25364_fsc.xml emd_25364_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_25364.png emd_25364.png | 137.1 KB | ||
| Filedesc metadata |  emd-25364.cif.gz emd-25364.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25364 http://ftp.pdbj.org/pub/emdb/structures/EMD-25364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25364 | HTTPS FTP |
-Related structure data
| Related structure data |  7sp1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11004 (Title: Cryo-EM structure of RNA-induced tau fibrils reveals a small C-terminal core that may nucleate fibril formation EMPIAR-11004 (Title: Cryo-EM structure of RNA-induced tau fibrils reveals a small C-terminal core that may nucleate fibril formationData size: 2.4 TB Data #1: Unaligned movies of recombinant tau amyloid fibrils formed in the presence of RNA [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25364.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25364.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Complex of tau protein and mouse RNA
| Entire | Name: Complex of tau protein and mouse RNA |
|---|---|
| Components |
|
-Supramolecule #1: Complex of tau protein and mouse RNA
| Supramolecule | Name: Complex of tau protein and mouse RNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform Tau-F of Microtubule-associated protein tau
| Macromolecule | Name: Isoform Tau-F of Microtubule-associated protein tau / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.919871 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKSTP TAEDVTAPLV DEGAPGKQA AAQPHTEIPE GTTAEEAGIG DTPSLEDEAA GHVTQARMVS KSKDGTGSDD KKAKGADGKT KIATPRGAAP P GQKGQANA ...String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKSTP TAEDVTAPLV DEGAPGKQA AAQPHTEIPE GTTAEEAGIG DTPSLEDEAA GHVTQARMVS KSKDGTGSDD KKAKGADGKT KIATPRGAAP P GQKGQANA TRIPAKTPPA PKTPPSSGEP PKSGDRSGYS SPGSPGTPGS RSRTPSLPTP PTREPKKVAV VRTPPKSPSS AK SRLQTAP VPMPDLKNVK SKIGSTENLK HQPGGGKVQI INKKLDLSNV QSKCGSKDNI KHVPGGGSVQ IVYKPVDLSK VTS KCGSLG NIHHKPGGGQ VEVKSEKLDF KDRVQSKIGS LDNITHVPGG GNKKIETHKL TFRENAKAKT DHGAEIVYKS PVVS GDTSP RHLSNVSSTG SIDMVDSPQL ATLADEVSAS LAKQGL UniProtKB: Isoform Tau-F of Microtubule-associated protein tau |
-Macromolecule #2: RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3')
| Macromolecule | Name: RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') / type: rna / ID: 2 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 3.2471 KDa |
| Sequence | String: AAAAAAAAAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Concentration: 20.0 mM / Component - Name: ammonium acetate |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 240 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 4728 / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.9 µm / Nominal defocus min: 0.76 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 84.4 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-7sp1: |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)