[English] 日本語
 Yorodumi
Yorodumi- EMDB-24899: CryoEM structure of Gi-coupled MRGPRX2 with small molecule agonis... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24899 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Gi-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573 | |||||||||
 Map data Map data | Deep sharp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmast cell secretagogue receptor activity / mast cell activation / sleep / neuropeptide binding / mast cell degranulation / positive regulation of cytokinesis / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway ...mast cell secretagogue receptor activity / mast cell activation / sleep / neuropeptide binding / mast cell degranulation / positive regulation of cytokinesis / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / sensory perception of pain / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / electron transport chain / negative regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / iron ion binding / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / heme binding / synapse / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
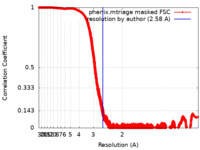
| Method | single particle reconstruction / cryo EM / Resolution: 2.58 Å | |||||||||
 Authors Authors | Cao C / Fay JF | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structure, function and pharmacology of human itch GPCRs. Authors: Can Cao / Hye Jin Kang / Isha Singh / He Chen / Chengwei Zhang / Wenlei Ye / Byron W Hayes / Jing Liu / Ryan H Gumpper / Brian J Bender / Samuel T Slocum / Brian E Krumm / Katherine Lansu / ...Authors: Can Cao / Hye Jin Kang / Isha Singh / He Chen / Chengwei Zhang / Wenlei Ye / Byron W Hayes / Jing Liu / Ryan H Gumpper / Brian J Bender / Samuel T Slocum / Brian E Krumm / Katherine Lansu / John D McCorvy / Wesley K Kroeze / Justin G English / Jeffrey F DiBerto / Reid H J Olsen / Xi-Ping Huang / Shicheng Zhang / Yongfeng Liu / Kuglae Kim / Joel Karpiak / Lily Y Jan / Soman N Abraham / Jian Jin / Brian K Shoichet / Jonathan F Fay / Bryan L Roth /   Abstract: The MRGPRX family of receptors (MRGPRX1-4) is a family of mas-related G-protein-coupled receptors that have evolved relatively recently. Of these, MRGPRX2 and MRGPRX4 are key physiological and ...The MRGPRX family of receptors (MRGPRX1-4) is a family of mas-related G-protein-coupled receptors that have evolved relatively recently. Of these, MRGPRX2 and MRGPRX4 are key physiological and pathological mediators of itch and related mast cell-mediated hypersensitivity reactions. MRGPRX2 couples to both G and G in mast cells. Here we describe agonist-stabilized structures of MRGPRX2 coupled to G and G in ternary complexes with the endogenous peptide cortistatin-14 and with a synthetic agonist probe, respectively, and the development of potent antagonist probes for MRGPRX2. We also describe a specific MRGPRX4 agonist and the structure of this agonist in a complex with MRGPRX4 and G. Together, these findings should accelerate the structure-guided discovery of therapeutic agents for pain, itch and mast cell-mediated hypersensitivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24899.map.gz emd_24899.map.gz | 54.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24899-v30.xml emd-24899-v30.xml emd-24899.xml emd-24899.xml | 27.4 KB 27.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24899_fsc.xml emd_24899_fsc.xml | 6.1 MB | Display |  FSC data file FSC data file |
| Images |  emd_24899.png emd_24899.png | 135 KB | ||
| Masks |  emd_24899_msk_1.map emd_24899_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24899.cif.gz emd-24899.cif.gz | 7.9 KB | ||
| Others |  emd_24899_additional_1.map.gz emd_24899_additional_1.map.gz emd_24899_half_map_1.map.gz emd_24899_half_map_1.map.gz emd_24899_half_map_2.map.gz emd_24899_half_map_2.map.gz | 59.5 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24899 http://ftp.pdbj.org/pub/emdb/structures/EMD-24899 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24899 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24899 | HTTPS FTP |
-Related structure data
| Related structure data |  7s8oMC  7s8lC  7s8mC  7s8nC  7s8pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10856 (Title: cryoEM structure of Gi-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573 EMPIAR-10856 (Title: cryoEM structure of Gi-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573Data size: 1.7 TB Data #1: Unaligned multi-frame micrographs [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24899.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24899.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Deep sharp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24899_msk_1.map emd_24899_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
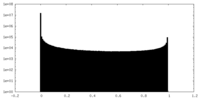
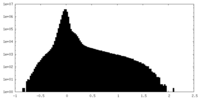
| Density Histograms |
-Additional map: sharp map
| File | emd_24899_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharp map | ||||||||||||
| Projections & Slices |
| ||||||||||||
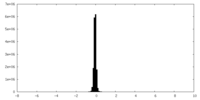
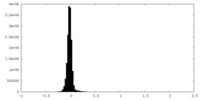
| Density Histograms |
-Half map: half map
| File | emd_24899_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
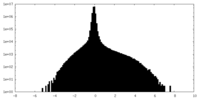
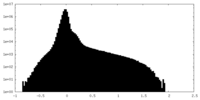
| Density Histograms |
-Half map: half map
| File | emd_24899_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
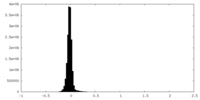
| Density Histograms |
- Sample components
Sample components
-Entire : MRGPRX2-Gi1 (R)-ZINC-3573
| Entire | Name: MRGPRX2-Gi1 (R)-ZINC-3573 |
|---|---|
| Components |
|
-Supramolecule #1: MRGPRX2-Gi1 (R)-ZINC-3573
| Supramolecule | Name: MRGPRX2-Gi1 (R)-ZINC-3573 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 kDa/nm |
-Macromolecule #1: Soluble cytochrome b562,Mas-related G-protein coupled receptor me...
| Macromolecule | Name: Soluble cytochrome b562,Mas-related G-protein coupled receptor member X2 chimera type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.054625 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DYKDDDDAKL QTMHHHHHHH HHHENLYFQG GTTMADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LGSTLEVLFQ GPDPTTPAWG T ESTTVNGN ...String: DYKDDDDAKL QTMHHHHHHH HHHENLYFQG GTTMADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LGSTLEVLFQ GPDPTTPAWG T ESTTVNGN DQALLLLCGK ETLIPVFLIL FIALVGLVGN GFVLWLLGFR MRRNAFSVYV LSLAGADFLF LCFQIINCLV YL SNFFCSI SINFPSFFTT VMTCAYLAGL SMLSTVSTER CLSVLWPIWY RCRRPRHLSA VVCVLLWALS LLLSILEGKF CGF LFSDGD SGWCQTFDFI TAAWLIFLFM VLCGSSLALL VRILCGSRGL PLTRLYLTIL LTVLVFLLCG LPFGIQWFLI LWIW KDSDV LFCHIHPVSV VLSSLNSSAN PIIYFFVGSF RKQWRLQQPI LKLALQRALQ DIAEVDHSEG CFRQGTPEMS RSSLV UniProtKB: Soluble cytochrome b562, Mas-related G-protein coupled receptor member X2 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.668922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAALEVLFQ GPHHHHHHHH |
-Macromolecule #6: (3R)-N,N-dimethyl-1-[(8S)-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl]p...
| Macromolecule | Name: (3R)-N,N-dimethyl-1-[(8S)-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl]pyrrolidin-3-amine type: ligand / ID: 6 / Number of copies: 1 / Formula: 8IU |
|---|---|
| Molecular weight | Theoretical: 307.393 Da |
| Chemical component information | 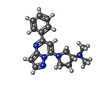 ChemComp-8IU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)