[English] 日本語
 Yorodumi
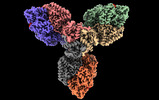
Yorodumi- EMDB-17454: Single particle cryo-EM structure of homohexameric 2-oxoglutarate... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum in complex with the product succinyl-CoA | |||||||||
 Map data Map data | OdhA_SCA primary map (no density mod) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | 2-oxoglutarate dehydrogenase / ODH / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationoxoglutarate dehydrogenase (succinyl-transferring) / oxoglutarate dehydrogenase (succinyl-transferring) activity / dihydrolipoyllysine-residue succinyltransferase / dihydrolipoyllysine-residue succinyltransferase activity / oxoglutarate dehydrogenase complex / thiamine pyrophosphate binding / tricarboxylic acid cycle / magnesium ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Corynebacterium glutamicum ATCC 13032 (bacteria) Corynebacterium glutamicum ATCC 13032 (bacteria) | |||||||||
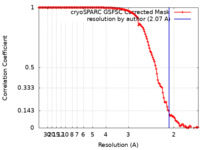
| Method | single particle reconstruction / cryo EM / Resolution: 2.07 Å | |||||||||
 Authors Authors | Yang L / Mechaly AM / Bellinzoni M | |||||||||
| Funding support |  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase. Authors: Lu Yang / Tristan Wagner / Ariel Mechaly / Alexandra Boyko / Eduardo M Bruch / Daniela Megrian / Francesca Gubellini / Pedro M Alzari / Marco Bellinzoni /     Abstract: Actinobacteria possess unique ways to regulate the oxoglutarate metabolic node. Contrary to most organisms in which three enzymes compose the 2-oxoglutarate dehydrogenase complex (ODH), ...Actinobacteria possess unique ways to regulate the oxoglutarate metabolic node. Contrary to most organisms in which three enzymes compose the 2-oxoglutarate dehydrogenase complex (ODH), actinobacteria rely on a two-in-one protein (OdhA) in which both the oxidative decarboxylation and succinyl transferase steps are carried out by the same polypeptide. Here we describe high-resolution cryo-EM and crystallographic snapshots of representative enzymes from Mycobacterium smegmatis and Corynebacterium glutamicum, showing that OdhA is an 800-kDa homohexamer that assembles into a three-blade propeller shape. The obligate trimeric and dimeric states of the acyltransferase and dehydrogenase domains, respectively, are critical for maintaining the overall assembly, where both domains interact via subtle readjustments of their interfaces. Complexes obtained with substrate analogues, reaction products and allosteric regulators illustrate how these domains operate. Furthermore, we provide additional insights into the phosphorylation-dependent regulation of this enzymatic machinery by the signalling protein OdhI. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: High resolution cryo-EM and crystallographic snapshots of the large actinobacterial 2-oxoglutarate dehydrogenase: an all-in-one fusion with unique properties Authors: Yang L / Wagner T / Mechaly A / Boyko A / Bruch EM / Megrian D / Gubellini F / Alzari PM / Bellinzoni M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17454.map.gz emd_17454.map.gz | 100.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17454-v30.xml emd-17454-v30.xml emd-17454.xml emd-17454.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17454_fsc.xml emd_17454_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17454.png emd_17454.png | 77.3 KB | ||
| Filedesc metadata |  emd-17454.cif.gz emd-17454.cif.gz | 7.7 KB | ||
| Others |  emd_17454_half_map_1.map.gz emd_17454_half_map_1.map.gz emd_17454_half_map_2.map.gz emd_17454_half_map_2.map.gz | 198.4 MB 198.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17454 http://ftp.pdbj.org/pub/emdb/structures/EMD-17454 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17454 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17454 | HTTPS FTP |
-Related structure data
| Related structure data |  8p5vMC  8p5rC  8p5sC  8p5tC  8p5uC  8p5wC  8p5xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17454.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17454.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | OdhA_SCA primary map (no density mod) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: OdhA SCA half map A cryoSPARC
| File | emd_17454_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | OdhA_SCA_half_map_A_cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: OdhA SCA half map B cryoSPARC
| File | emd_17454_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | OdhA_SCA_half_map_B_cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homohexameric 2-oxoglutarate dehydrogenase
| Entire | Name: Homohexameric 2-oxoglutarate dehydrogenase |
|---|---|
| Components |
|
-Supramolecule #1: Homohexameric 2-oxoglutarate dehydrogenase
| Supramolecule | Name: Homohexameric 2-oxoglutarate dehydrogenase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum ATCC 13032 (bacteria) Corynebacterium glutamicum ATCC 13032 (bacteria) |
| Molecular weight | Theoretical: 808 KDa |
-Macromolecule #1: 2-oxoglutarate dehydrogenase E1/E2 component
| Macromolecule | Name: 2-oxoglutarate dehydrogenase E1/E2 component / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO EC number: oxoglutarate dehydrogenase (succinyl-transferring) |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum ATCC 13032 (bacteria) Corynebacterium glutamicum ATCC 13032 (bacteria) |
| Molecular weight | Theoretical: 134.972078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMSSASTFG QNAWLVDEMF QQFQKDPKSV DKEWRELFEA QGGPNTTPAT TEAQPSAPKE SAKPAPKAAP AAKAAPRVET KPADKTAPK AKESSVPQQP KLPEPGQTPI RGIFKSIAKN MDISLEIPTA TSVRDMPARL MFENRAMVND QLKRTRGGKI S FTHIIGYA ...String: GSMSSASTFG QNAWLVDEMF QQFQKDPKSV DKEWRELFEA QGGPNTTPAT TEAQPSAPKE SAKPAPKAAP AAKAAPRVET KPADKTAPK AKESSVPQQP KLPEPGQTPI RGIFKSIAKN MDISLEIPTA TSVRDMPARL MFENRAMVND QLKRTRGGKI S FTHIIGYA MVKAVMAHPD MNNSYDVIDG KPTLIVPEHI NLGLAIDLPQ KDGSRALVVA AIKETEKMNF SEFLAAYEDI VA RSRKGKL TMDDYQGVTV SLTNPGGIGT RHSVPRLTKG QGTIIGVGSM DYPAEFQGAS EDRLAELGVG KLVTITSTYD HRV IQGAVS GEFLRTMSRL LTDDSFWDEI FDAMNVPYTP MRWAQDVPNT GVDKNTRVMQ LIEAYRSRGH LIADTNPLSW VQPG MPVPD HRDLDIETHN LTIWDLDRTF NVGGFGGKET MTLREVLSRL RAAYTLKVGS EYTHILDRDE RTWLQDRLEA GMPKP TQAE QKYILQKLNA AEAFENFLQT KYVGQKRFSL EGAEALIPLM DSAIDTAAGQ GLDEVVIGMP HRGRLNVLFN IVGKPL ASI FNEFEGQMEQ GQIGGSGDVK YHLGSEGQHL QMFGDGEIKV SLTANPSHLE AVNPVMEGIV RAKQDYLDKG VDGKTVV PL LLHGDAAFAG LGIVPETINL AKLRGYDVGG TIHIVVNNQI GFTTTPDSSR SMHYATDYAK AFGCPVFHVN GDDPEAVV W VGQLATEYRR RFGKDVFIDL VCYRLRGHNE ADDPSMTQPK MYELITGRET VRAQYTEDLL GRGDLSNEDA EAVVRDFHD QMESVFNEVK EGGKKQAEAQ TGITGSQKLP HGLETNISRE ELLELGQAFA NTPEGFNYHP RVAPVAKKRV SSVTEGGIDW AWGELLAFG SLANSGRLVR LAGEDSRRGT FTQRHAVAID PATAEEFNPL HELAQSKGNN GKFLVYNSAL TEYAGMGFEY G YSVGNEDS IVAWEAQFGD FANGAQTIID EYVSSGEAKW GQTSKLILLL PHGYEGQGPD HSSARIERFL QLCAEGSMTV AQ PSTPANH FHLLRRHALS DLKRPLVIFT PKSMLRNKAA ASAPEDFTEV TKFQSVINDP NVADAAKVKK VMLVSGKLYY ELA KRKEKD GRDDIAIVRI EMLHPIPFNR ISEALAGYPN AEEVLFVQDE PANQGPWPFY QEHLPELIPN MPKMRRVSRR AQSS TATGV AKVHQLEEKQ LIDEAFEA UniProtKB: 2-oxoglutarate dehydrogenase E1/E2 component |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: ACETYL COENZYME *A
| Macromolecule | Name: ACETYL COENZYME *A / type: ligand / ID: 3 / Number of copies: 6 / Formula: ACO |
|---|---|
| Molecular weight | Theoretical: 809.571 Da |
| Chemical component information |  ChemComp-ACO: |
-Macromolecule #4: THIAMINE DIPHOSPHATE
| Macromolecule | Name: THIAMINE DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 6 / Formula: TPP |
|---|---|
| Molecular weight | Theoretical: 425.314 Da |
| Chemical component information |  ChemComp-TPP: |
-Macromolecule #5: SUCCINYL-COENZYME A
| Macromolecule | Name: SUCCINYL-COENZYME A / type: ligand / ID: 5 / Number of copies: 6 / Formula: SCA |
|---|---|
| Molecular weight | Theoretical: 867.607 Da |
| Chemical component information |  ChemComp-SCA: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 1280 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 12.0 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 / Component:
| ||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 11827 / Average exposure time: 1.9 sec. / Average electron dose: 48.0 e/Å2 / Details: Each image was composed by 50 frames |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8p5v: |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)