[English] 日本語
 Yorodumi
Yorodumi- EMDB-16900: Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution | |||||||||
 Map data Map data | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dihydrolipoyl Succinyltransferase / E2 / Oxoglutarate / a-Ketoglutarate / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-lysine catabolic process to acetyl-CoA via L-saccharopine / dihydrolipoyllysine-residue succinyltransferase / dihydrolipoyllysine-residue succinyltransferase activity / oxoglutarate dehydrogenase complex / tricarboxylic acid cycle / mitochondrion Similarity search - Function | |||||||||
| Biological species |  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||
 Authors Authors | Skalidis I / Tueting C / Kyrilis FL / Hamdi F / Kastritis PL | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Structural analysis of an endogenous 4-megadalton succinyl-CoA-generating metabolon. Authors: Ioannis Skalidis / Fotis L Kyrilis / Christian Tüting / Farzad Hamdi / Toni K Träger / Jaydeep Belapure / Gerd Hause / Marta Fratini / Francis J O'Reilly / Ingo Heilmann / Juri Rappsilber ...Authors: Ioannis Skalidis / Fotis L Kyrilis / Christian Tüting / Farzad Hamdi / Toni K Träger / Jaydeep Belapure / Gerd Hause / Marta Fratini / Francis J O'Reilly / Ingo Heilmann / Juri Rappsilber / Panagiotis L Kastritis /     Abstract: The oxoglutarate dehydrogenase complex (OGDHc) participates in the tricarboxylic acid cycle and, in a multi-step reaction, decarboxylates α-ketoglutarate, transfers succinyl to CoA, and reduces NAD+. ...The oxoglutarate dehydrogenase complex (OGDHc) participates in the tricarboxylic acid cycle and, in a multi-step reaction, decarboxylates α-ketoglutarate, transfers succinyl to CoA, and reduces NAD+. Due to its pivotal role in metabolism, OGDHc enzymatic components have been studied in isolation; however, their interactions within the endogenous OGDHc remain elusive. Here, we discern the organization of a thermophilic, eukaryotic, native OGDHc in its active state. By combining biochemical, biophysical, and bioinformatic methods, we resolve its composition, 3D architecture, and molecular function at 3.35 Å resolution. We further report the high-resolution cryo-EM structure of the OGDHc core (E2o), which displays various structural adaptations. These include hydrogen bonding patterns confining interactions of OGDHc participating enzymes (E1o-E2o-E3), electrostatic tunneling that drives inter-subunit communication, and the presence of a flexible subunit (E3BPo), connecting E2o and E3. This multi-scale analysis of a succinyl-CoA-producing native cell extract provides a blueprint for structure-function studies of complex mixtures of medical and biotechnological value. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16900.map.gz emd_16900.map.gz | 32.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16900-v30.xml emd-16900-v30.xml emd-16900.xml emd-16900.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
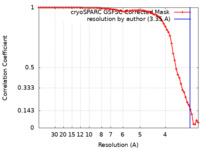
| FSC (resolution estimation) |  emd_16900_fsc.xml emd_16900_fsc.xml | 7.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_16900.png emd_16900.png | 84.2 KB | ||
| Masks |  emd_16900_msk_1.map emd_16900_msk_1.map | 34.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16900.cif.gz emd-16900.cif.gz | 6.3 KB | ||
| Others |  emd_16900_additional_1.map.gz emd_16900_additional_1.map.gz emd_16900_half_map_1.map.gz emd_16900_half_map_1.map.gz emd_16900_half_map_2.map.gz emd_16900_half_map_2.map.gz | 2.5 MB 31.8 MB 31.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16900 http://ftp.pdbj.org/pub/emdb/structures/EMD-16900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16900 | HTTPS FTP |
-Related structure data
| Related structure data |  8oiuMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16900.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16900.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5678 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16900_msk_1.map emd_16900_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Cryo-EM reconstruction of the native asymmetric complex of...
| File | emd_16900_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the native asymmetric complex of the 2-oxoglutarate dehydrogenase complex of C. thermophilum. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM reconstruction of the native 24-mer E2o core...
| File | emd_16900_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum, Half-Map A. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM reconstruction of the native 24-mer E2o core...
| File | emd_16900_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum, Half-Map B. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Native 24-mer core of Oxoglutarate Dehydrogenase Complex
| Entire | Name: Native 24-mer core of Oxoglutarate Dehydrogenase Complex |
|---|---|
| Components |
|
-Supramolecule #1: Native 24-mer core of Oxoglutarate Dehydrogenase Complex
| Supramolecule | Name: Native 24-mer core of Oxoglutarate Dehydrogenase Complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) |
| Molecular weight | Theoretical: 3 MDa |
-Macromolecule #1: Dihydrolipoyllysine-residue succinyltransferase
| Macromolecule | Name: Dihydrolipoyllysine-residue succinyltransferase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: dihydrolipoyllysine-residue succinyltransferase |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) |
| Molecular weight | Theoretical: 45.987004 KDa |
| Sequence | String: MLYRGLRMAA RAAPKFAVLN THAALRQLPL QFQHVRTYAD KIIKVPQMAE SITEGTLKQW NKAVGDYVEA DEEIATIETD KIDVAVNAP EAGVIKEFFV NEEDTVLVGQ DLVRLEVGGE KPAEAAKEQP KAAAPEPKVE EKKVPEAPAP EPSKTAAPAP A PPKQEAPA ...String: MLYRGLRMAA RAAPKFAVLN THAALRQLPL QFQHVRTYAD KIIKVPQMAE SITEGTLKQW NKAVGDYVEA DEEIATIETD KIDVAVNAP EAGVIKEFFV NEEDTVLVGQ DLVRLEVGGE KPAEAAKEQP KAAAPEPKVE EKKVPEAPAP EPSKTAAPAP A PPKQEAPA SPKPASKPAE TPAVTLGNRE ERRVKMNRMR LRIAERLKQS QNTAASLTTF NEVDMSALIE FRNKYKDEVL KK TGVKLGF MSAFSRAVVL AIRDLPVVNA SIEGPNGGDT IVYRDYVDIS VAVATEKGLV TPVVRNAETM DLITIEKTIA ELG KKARDG KLTIEDMAGG TFTISNGGVF GSLMGTPIIN LPQSAVLGLH AIKERPVAVN GKVEIRPMMY LALTYDHRLL DGRE AVQFL VKVKEYIEDP RKMLL UniProtKB: dihydrolipoyllysine-residue succinyltransferase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 200.0 mM / Component - Formula: NH4CH2COOH / Component - Name: Ammonium acetate |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: For plunging, blot force 0 and blotting time of 4 sec were applied.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Temperature | Min: 77.15 K / Max: 103.15 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 21381 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 92000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: all / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Initial fitting was performed in ChimeraX v1.2, then real-space refined in PHENIX v1.19 |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8oiu: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)