[English] 日本語
 Yorodumi
Yorodumi- EMDB-16572: 1.85 angstrom resolution cryo-EM reconstruction of tobacco mosaic... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1.85 angstrom resolution cryo-EM reconstruction of tobacco mosaic virus, imaged on a JEOL CryoARM 300 with Direct Electron Apollo camera. | |||||||||
 Map data Map data | Reconstruction of tobacco mosaic virus | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Tobacco mosaic virus Tobacco mosaic virus | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 1.85 Å | |||||||||
 Authors Authors | Bhella D / Love AJ / Streetley J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: 1.85 angstrom resolution cryo-EM reconstruction of tobacco mosaic virus, imaged on a JEOL CryoARM 300 with Direct Electron Apollo camera. Authors: Bhella D / Love AJ / Streetley J / Taliansky M / McGeachy K / Bukharova T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16572.map.gz emd_16572.map.gz | 161.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16572-v30.xml emd-16572-v30.xml emd-16572.xml emd-16572.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
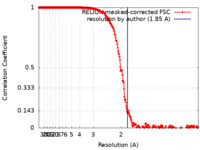
| FSC (resolution estimation) |  emd_16572_fsc.xml emd_16572_fsc.xml | 26.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_16572.png emd_16572.png | 175.5 KB | ||
| Others |  emd_16572_half_map_1.map.gz emd_16572_half_map_1.map.gz emd_16572_half_map_2.map.gz emd_16572_half_map_2.map.gz | 1.3 GB 1.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16572 http://ftp.pdbj.org/pub/emdb/structures/EMD-16572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16572 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16572.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16572.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of tobacco mosaic virus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.516 Å | ||||||||||||||||||||||||||||||||||||
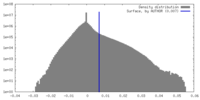
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Reconstruction of tobacco mosaic virus Half map 1
| File | emd_16572_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of tobacco mosaic virus Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
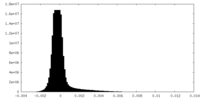
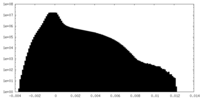
| Density Histograms |
-Half map: Reconstruction of tobacco mosaic virus Half map 2
| File | emd_16572_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of tobacco mosaic virus Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
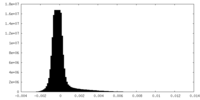
| Density Histograms |
- Sample components
Sample components
-Entire : Tobacco mosaic virus
| Entire | Name:   Tobacco mosaic virus Tobacco mosaic virus |
|---|---|
| Components |
|
-Supramolecule #1: Tobacco mosaic virus
| Supramolecule | Name: Tobacco mosaic virus / type: virus / ID: 1 / Parent: 0 Details: Infected leaf material was ground in phosphate buffer, cleared with chloroform and centrifuged (4 degrees C, 10000g) for 20mins. The supernatant was then precipitated overnight at 4 degrees ...Details: Infected leaf material was ground in phosphate buffer, cleared with chloroform and centrifuged (4 degrees C, 10000g) for 20mins. The supernatant was then precipitated overnight at 4 degrees C after adding both NaCl and PEG8000 to 1% and 2% (w/v) respectively. This was centrifuged at 4 degrees C 10000g for 20 mins, and pellets were resuspended in 25 mM Tris-HCl pH 7.8, prior to loading onto 20% sucrose cushions made up in this buffer. The cushions were centrifuged at 175,500g at 4 degrees C for 2 h and the subsequently resuspended in 0.01 M Tris-HCl buffer (pH 7.8) or water prior to centrifugation at 175,500g at 4 degrees C for 2 h. The pellets were resuspended and centrifuged again as before. The pellets were finally reconstituted in water with 0.02% sodium azide. NCBI-ID: 12242 / Sci species name: Tobacco mosaic virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 24 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: Not buffered - sample was in H2O |
| Grid | Model: C-flat / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 30 eV |
| Image recording | Film or detector model: DIRECT ELECTRON APOLLO (4k x 4k) / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Average electron dose: 60.0 e/Å2 Details: Images collected in super-resolution mode (0.387 angstroms/pixel). Calibrated pixel size 0.774 angstroms/pixel. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)