+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Neurofascin isoform NF155 extracellular domain | |||||||||
 Map data Map data | Volume map of the NF155 extracellular domain obtained by negative stain EM. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
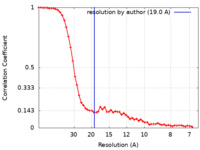
| Method | single particle reconstruction / negative staining / Resolution: 19.0 Å | |||||||||
 Authors Authors | McKie SJ / Deane JE / Butt BG | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Altered plasma membrane abundance of the sulfatide-binding protein NF155 links glycosphingolipid imbalances to demyelination. Authors: Shannon J McKie / Alex S Nicholson / Emily Smith / Stuart Fawke / Eve R Caroe / James C Williamson / Benjamin G Butt / Denisa Kolářová / Ondřej Peterka / Michal Holčapek / Paul J Lehner ...Authors: Shannon J McKie / Alex S Nicholson / Emily Smith / Stuart Fawke / Eve R Caroe / James C Williamson / Benjamin G Butt / Denisa Kolářová / Ondřej Peterka / Michal Holčapek / Paul J Lehner / Stephen C Graham / Janet E Deane /   Abstract: Myelin is a multilayered membrane that tightly wraps neuronal axons, enabling efficient, high-speed signal propagation. The axon and myelin sheath form tight contacts, mediated by specific plasma ...Myelin is a multilayered membrane that tightly wraps neuronal axons, enabling efficient, high-speed signal propagation. The axon and myelin sheath form tight contacts, mediated by specific plasma membrane proteins and lipids, and disruption of these contacts causes devastating demyelinating diseases. Using two cell-based models of demyelinating sphingolipidoses, we demonstrate that altered lipid metabolism changes the abundance of specific plasma membrane proteins. These altered membrane proteins have known roles in cell adhesion and signaling, with several implicated in neurological diseases. The cell surface abundance of the adhesion molecule neurofascin (NFASC), a protein critical for the maintenance of myelin-axon contacts, changes following disruption to sphingolipid metabolism. This provides a direct molecular link between altered lipid abundance and myelin stability. We show that the NFASC isoform NF155, but not NF186, interacts directly and specifically with the sphingolipid sulfatide via multiple binding sites and that this interaction requires the full-length extracellular domain of NF155. We demonstrate that NF155 adopts an S-shaped conformation and preferentially binds sulfatide-containing membranes in , with important implications for protein arrangement in the tight axon-myelin space. Our work links glycosphingolipid imbalances to disturbance of membrane protein abundance and demonstrates how this may be driven by direct protein-lipid interactions, providing a mechanistic framework to understand the pathogenesis of galactosphingolipidoses. #1:  Journal: BioRxiv / Year: 2022 Journal: BioRxiv / Year: 2022Title: Altered plasma membrane abundance of the sulfatide-binding protein NF155 links glycosphingolipid imbalances to demyelination Authors: McKie SJ / Deane JE / Nicholson A / Smith E / Fawke S / Caroe E / Williamson JC / Butt BG / Kolarova D / Peterka O / Holcapek M / Lehner PJ / Graham SC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16540.map.gz emd_16540.map.gz | 6.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16540-v30.xml emd-16540-v30.xml emd-16540.xml emd-16540.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16540_fsc.xml emd_16540_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16540.png emd_16540.png | 47.6 KB | ||
| Others |  emd_16540_half_map_1.map.gz emd_16540_half_map_1.map.gz emd_16540_half_map_2.map.gz emd_16540_half_map_2.map.gz | 12 MB 12 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16540 http://ftp.pdbj.org/pub/emdb/structures/EMD-16540 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16540 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16540 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16540.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16540.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Volume map of the NF155 extracellular domain obtained by negative stain EM. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.31 Å | ||||||||||||||||||||||||||||||||||||
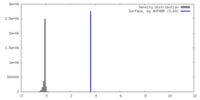
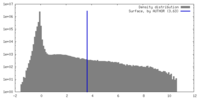
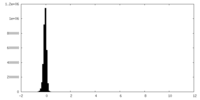
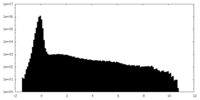
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EM half map of the NF155 extracellular domain...
| File | emd_16540_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map of the NF155 extracellular domain obtained by negative stain EM. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EM half map of the NF155 extracellular domain...
| File | emd_16540_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map of the NF155 extracellular domain obtained by negative stain EM. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Neurofascin isoform NF155
| Entire | Name: Neurofascin isoform NF155 |
|---|---|
| Components |
|
-Supramolecule #1: Neurofascin isoform NF155
| Supramolecule | Name: Neurofascin isoform NF155 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Full length extracellular domain of neurofascin isoform NF155 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: Neurofascin isoform NF155
| Macromolecule | Name: Neurofascin isoform NF155 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IEIPMDLTQP PTITKQSAKD HIVDPRDNIL IECEAKGNPA PSFHWTRNSR FFNIAKDPRV SMRRRSGTLV IDFRSGGRPE EYEGEYQCFA RNKFGTALSN RIRLQVSKSP LWPKENLDPV VVQEGAPLTL QCNPPPGLPS PVIFWMSSSM EPITQDKRVS QGHNGDLYFS ...String: IEIPMDLTQP PTITKQSAKD HIVDPRDNIL IECEAKGNPA PSFHWTRNSR FFNIAKDPRV SMRRRSGTLV IDFRSGGRPE EYEGEYQCFA RNKFGTALSN RIRLQVSKSP LWPKENLDPV VVQEGAPLTL QCNPPPGLPS PVIFWMSSSM EPITQDKRVS QGHNGDLYFS NVMLQDMQTD YSCNARFHFT HTIQQKNPFT LKVLTNHPYN DSSLRNHPDM YSARGVAERT PSFMYPQGTA SSQMVLRGMD LLLECIASGV PTPDIAWYKK GGDLPSDKAK FENFNKALRI TNVSEEDSGE YFCLASNKMG SIRHTISVRV KAAPYWLDEP KNLILAPGED GRLVCRANGN PKPTVQWMVN GEPLQSAPPN PNREVAGDTI IFRDTQISSR AVYQCNTSNE HGYLLANAFV SVLDVPPRML SPRNQLIRVI LYNRTRLDCP FFGSPIPTLR WFKNGQGSNL DGGNYHVYEN GSLEIKMIRK EDQGIYTCVA TNILGKAENQ VRLEVKDPTR IYRMPEDQVA RRGTTVQLEC RVKHDPSLKL TVSWLKDDEP LYIGNRMKKE DDSLTIFGVA ERDQGSYTCV ASTELDQDLA KAYLTVLGRP DRPRDLELTD LAERSVRLTW IPGDANNSPI TDYVVQFEED QFQPGVWHDH SKYPGSVNSA VLRLSPYVNY QFRVIAINEV GSSHPSLPSE RYRTSGAPPE SNPGDVKGEG TRKNNMEITW TPMNATSAFG PNLRYIVKWR RRETREAWNN VTVWGSRYVV GQTPVYVPYE IRVQAENDFG KGPEPESVIG YSGEDYPRAA PTEVKVRVMN STAISLQWNR VYSDTVQGQL REYRAYYWRE SSLLKNLWVS QKRQQASFPG DRLRGVVSRL FPYSNYKLEM VVVNGRGDGP RSETKEFTTP EGVPSAPRRF RVRQPNLETI NLEWDHPEHP NGIMIGYTLK YVAFNGTKVG KQIVENFSPN QTKFTVQRTD PVSRYRFTLS ARTQVGSGEA VTEESPAPPN EATPTAAYTN NQADIATQGK HHHHHH |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.0023 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
| Details | This sample was highly pure and monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)