[English] 日本語
 Yorodumi
Yorodumi- EMDB-16136: Cryo-electron tomogram acquired on a cryo-FIB lamella of a retina... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomogram acquired on a cryo-FIB lamella of a retinal pigment epithelial (RPE1) cell | |||||||||
 Map data Map data | Reconstructed cryo-electron tomogram acquired on RPE1 cryo-FIB lamella | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Actin stress fiber / CYTOSOLIC PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Mahamid J / Goetz SK | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation | Journal: Nat Methods / Year: 2020 Title: Tailoring cryo-electron microscopy grids by photo-micropatterning for in-cell structural studies. Authors: Mauricio Toro-Nahuelpan / Ievgeniia Zagoriy / Fabrice Senger / Laurent Blanchoin / Manuel Théry / Julia Mahamid /   Abstract: Spatially controlled cell adhesion on electron microscopy supports remains a bottleneck in specimen preparation for cellular cryo-electron tomography. Here, we describe contactless and mask-free ...Spatially controlled cell adhesion on electron microscopy supports remains a bottleneck in specimen preparation for cellular cryo-electron tomography. Here, we describe contactless and mask-free photo-micropatterning of electron microscopy grids for site-specific deposition of extracellular matrix-related proteins. We attained refined cell positioning for micromachining by cryo-focused ion beam milling. Complex micropatterns generated predictable intracellular organization, allowing direct correlation between cell architecture and in-cell three-dimensional structural characterization of the underlying molecular machinery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16136.map.gz emd_16136.map.gz | 361.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16136-v30.xml emd-16136-v30.xml emd-16136.xml emd-16136.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16136.png emd_16136.png | 342.6 KB | ||
| Masks |  emd_16136_msk_1.map emd_16136_msk_1.map | 2 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-16136.cif.gz emd-16136.cif.gz | 4.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16136 http://ftp.pdbj.org/pub/emdb/structures/EMD-16136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16136 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16136.map.gz / Format: CCP4 / Size: 513.3 MB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_16136.map.gz / Format: CCP4 / Size: 513.3 MB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstructed cryo-electron tomogram acquired on RPE1 cryo-FIB lamella | ||||||||||||||||||||
| Voxel size | X=Y=Z: 13.481 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16136_msk_1.map emd_16136_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
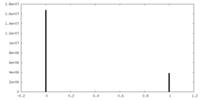
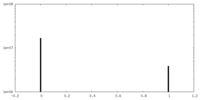
| Density Histograms |
- Sample components
Sample components
-Entire : RPE1 cytosol with actin stress fiber
| Entire | Name: RPE1 cytosol with actin stress fiber |
|---|---|
| Components |
|
-Supramolecule #1: RPE1 cytosol with actin stress fiber
| Supramolecule | Name: RPE1 cytosol with actin stress fiber / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: RPE1 cell line / Location in cell: cytosol Homo sapiens (human) / Strain: RPE1 cell line / Location in cell: cytosol |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil / Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE |
| Cryo protectant | None |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 / Focused ion beam - Current: 0.05 / Focused ion beam - Duration: 120 / Focused ion beam - Temperature: 88 K / Focused ion beam - Initial thickness: 1000 / Focused ion beam - Final thickness: 200 Focused ion beam - Details: The value given for _em_focused_ion_beam.instrument is Thermo Fisher Aquilos. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 42000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 4.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 57 IMOD / Number images used: 57 |
|---|
 Movie
Movie Controller
Controller






















 Z
Z Y
Y X
X