[English] 日本語
 Yorodumi
Yorodumi- EMDB-15735: Microtubule decorated with kinesin-motor domains, 14 protofilamen... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start helix, 1 seam | |||||||||||||||
 Map data Map data | Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start helix, 1 seam | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Cytoskeleton / Microtubule / Tubulin / kinesin / STRUCTURAL PROTEIN | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 25.0 Å | |||||||||||||||
 Authors Authors | Chretien D / Guyomar C | |||||||||||||||
| Funding support |  France, France,  Switzerland, 4 items Switzerland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Changes in seam number and location induce holes within microtubules assembled from porcine brain tubulin and in egg cytoplasmic extracts. Authors: Charlotte Guyomar / Clément Bousquet / Siou Ku / John M Heumann / Gabriel Guilloux / Natacha Gaillard / Claire Heichette / Laurence Duchesne / Michel O Steinmetz / Romain Gibeaux / Denis Chrétien /    Abstract: Microtubules are tubes of about 25 nm in diameter that are critically involved in a variety of cellular functions, including motility, compartmentalization, and division. They are considered as ...Microtubules are tubes of about 25 nm in diameter that are critically involved in a variety of cellular functions, including motility, compartmentalization, and division. They are considered as pseudo-helical polymers whose constituent αβ-tubulin heterodimers share lateral homotypic interactions, except at one unique region called the seam. Here, we used a segmented sub-tomogram averaging strategy to reassess this paradigm and analyze the organization of the αβ-tubulin heterodimers in microtubules assembled from purified porcine brain tubulin in the presence of GTP and GMPCPP, and in egg cytoplasmic extracts. We find that in almost all conditions, microtubules incorporate variable protofilament and/or tubulin subunit helical-start numbers, as well as variable numbers of seams. Strikingly, the seam number and location vary along individual microtubules, generating holes of one to a few subunits in size within their lattices. Together, our results reveal that the formation of mixed and discontinuous microtubule lattices is an intrinsic property of tubulin that requires the formation of unique lateral interactions without longitudinal ones. They further suggest that microtubule assembly is tightly regulated in a cytoplasmic environment. #1:  Journal: bioRxiv / Year: 2022 Journal: bioRxiv / Year: 2022Title: Changes in seam number and location induce holes within microtubules assembled from porcine brain tubulin and in Xenopus egg cytoplasmic extracts Authors: Guyomar C / Bousquet C / Ku S / Heumann J / Guilloux G / Gaillard N / Heichette C / Duchesne L / Steinmetz MO / Gibeaux R / Chretien D | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15735.map.gz emd_15735.map.gz | 857.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15735-v30.xml emd-15735-v30.xml emd-15735.xml emd-15735.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15735.png emd_15735.png | 104.5 KB | ||
| Filedesc metadata |  emd-15735.cif.gz emd-15735.cif.gz | 5.3 KB | ||
| Others |  emd_15735_additional_1.map.gz emd_15735_additional_1.map.gz emd_15735_half_map_1.map.gz emd_15735_half_map_1.map.gz emd_15735_half_map_2.map.gz emd_15735_half_map_2.map.gz | 887.4 KB 943 KB 943 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15735 http://ftp.pdbj.org/pub/emdb/structures/EMD-15735 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15735 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15735 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15735.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15735.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start helix, 1 seam | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.88 Å | ||||||||||||||||||||||||||||||||||||
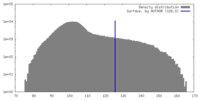
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start...
| File | emd_15735_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start helix, 1 seam. Rotated. | ||||||||||||
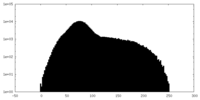
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start...
| File | emd_15735_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start helix, 1 seam, even map | ||||||||||||
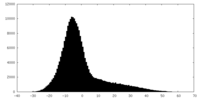
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start...
| File | emd_15735_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Microtubule decorated with kinesin-motor domains, 14 protofilaments, 3-start helix, 1 seam, odd map | ||||||||||||
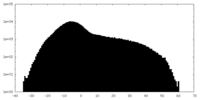
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Microtubule assembled from purified porcine brain alpha-beta tubu...
| Entire | Name: Microtubule assembled from purified porcine brain alpha-beta tubulin and decorated with kinesin-motor domain Kif5B |
|---|---|
| Components |
|
-Supramolecule #1: Microtubule assembled from purified porcine brain alpha-beta tubu...
| Supramolecule | Name: Microtubule assembled from purified porcine brain alpha-beta tubulin and decorated with kinesin-motor domain Kif5B type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Details: Microtubule assembled in the presence of GTP |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Kinesin-motor domain Kif5B
| Supramolecule | Name: Kinesin-motor domain Kif5B / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.8 Component:
Details: pH adjusted with KOH | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 308.15 K / Instrument: LEICA EM GP Details: Blot for 2 seconds using Whatman paper number 1 before plunging. | |||||||||||||||
| Details | Tubulin was mixed with 10 nm gold nanoparticles at 80 nM final concentration. The sample was incubated at 35 degrees for 1 hour. Kinesin-motor domains mixed with gold nanoparticles were added to the sample right before freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Temperature | Min: 88.15 K / Max: 93.15 K |
| Details | Tilt series were started at 0 degrees and acquired using a Saxton scheme |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Average exposure time: 1.0 sec. / Average electron dose: 1.0 e/Å2 Details: Images were acquired in binning 2 Electron dose was not calibrated |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: GATAN CT3500TR SINGLE TILT ROTATION LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PEET Details: The picking method involves a large overlap between sub-tomograms. The value provided by the FSC must be taken with caution (no Gold Standard). No FCS curve provided. Number subtomograms used: 178 |
|---|---|
| Extraction | Number tomograms: 1 / Number images used: 178 |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)