[English] 日本語
 Yorodumi
Yorodumi- EMDB-15533: Cryo-EM structure of the proline-rich antimicrobial peptide droso... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the 50S ribosomal subunit | |||||||||
 Map data Map data | post masked | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E.coli / Drosocin / Dro1 / antimicrobial peptide / proline-rich / PrAMP / 50S / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationassembly of large subunit precursor of preribosome / response to radiation / large ribosomal subunit / transferase activity / ribosome binding / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation ...assembly of large subunit precursor of preribosome / response to radiation / large ribosomal subunit / transferase activity / ribosome binding / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.1 Å | |||||||||
 Authors Authors | Koller TO / Morici M / Wilson DN | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Structural basis for translation inhibition by the glycosylated drosocin peptide. Authors: Timm O Koller / Martino Morici / Max Berger / Haaris A Safdari / Deepti S Lele / Bertrand Beckert / Kanwal J Kaur / Daniel N Wilson /    Abstract: The proline-rich antimicrobial peptide (PrAMP) drosocin is produced by Drosophila species to combat bacterial infection. Unlike many PrAMPs, drosocin is O-glycosylated at threonine 11, a post- ...The proline-rich antimicrobial peptide (PrAMP) drosocin is produced by Drosophila species to combat bacterial infection. Unlike many PrAMPs, drosocin is O-glycosylated at threonine 11, a post-translation modification that enhances its antimicrobial activity. Here we demonstrate that the O-glycosylation not only influences cellular uptake of the peptide but also interacts with its intracellular target, the ribosome. Cryogenic electron microscopy structures of glycosylated drosocin on the ribosome at 2.0-2.8-Å resolution reveal that the peptide interferes with translation termination by binding within the polypeptide exit tunnel and trapping RF1 on the ribosome, reminiscent of that reported for the PrAMP apidaecin. The glycosylation of drosocin enables multiple interactions with U2609 of the 23S rRNA, leading to conformational changes that break the canonical base pair with A752. Collectively, our study reveals novel molecular insights into the interaction of O-glycosylated drosocin with the ribosome, which provide a structural basis for future development of this class of antimicrobials. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15533.map.gz emd_15533.map.gz | 69 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15533-v30.xml emd-15533-v30.xml emd-15533.xml emd-15533.xml | 54.9 KB 54.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15533_fsc.xml emd_15533_fsc.xml | 20.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15533.png emd_15533.png | 104.9 KB | ||
| Filedesc metadata |  emd-15533.cif.gz emd-15533.cif.gz | 12 KB | ||
| Others |  emd_15533_half_map_1.map.gz emd_15533_half_map_1.map.gz emd_15533_half_map_2.map.gz emd_15533_half_map_2.map.gz | 672.6 MB 670.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15533 http://ftp.pdbj.org/pub/emdb/structures/EMD-15533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15533 | HTTPS FTP |
-Related structure data
| Related structure data |  8anaMC  8aknC  8am9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15533.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15533.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post masked | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8 Å | ||||||||||||||||||||||||||||||||||||
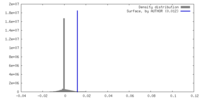
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half 2
| File | emd_15533_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
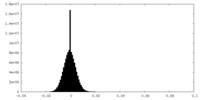
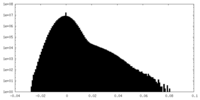
| Density Histograms |
-Half map: half 1
| File | emd_15533_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
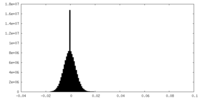
| Density Histograms |
- Sample components
Sample components
+Entire : Cryo-EM structure of the proline-rich antimicrobial peptide droso...
+Supramolecule #1: Cryo-EM structure of the proline-rich antimicrobial peptide droso...
+Macromolecule #1: 50S ribosomal protein L33
+Macromolecule #2: 50S ribosomal protein L34
+Macromolecule #3: 50S ribosomal protein L35
+Macromolecule #4: 50S ribosomal protein L36
+Macromolecule #5: 50S ribosomal protein L31
+Macromolecule #6: Drosocin1
+Macromolecule #9: 50S ribosomal protein L2
+Macromolecule #10: 50S ribosomal protein L3
+Macromolecule #11: 50S ribosomal protein L4
+Macromolecule #12: 50S ribosomal protein L5
+Macromolecule #13: 50S ribosomal protein L6
+Macromolecule #14: 50S ribosomal protein L9
+Macromolecule #15: 50S ribosomal protein L13
+Macromolecule #16: 50S ribosomal protein L14
+Macromolecule #17: 50S ribosomal protein L15
+Macromolecule #18: 50S ribosomal protein L16
+Macromolecule #19: 50S ribosomal protein L17
+Macromolecule #20: 50S ribosomal protein L18
+Macromolecule #21: 50S ribosomal protein L19
+Macromolecule #22: 50S ribosomal protein L20
+Macromolecule #23: 50S ribosomal protein L21
+Macromolecule #24: 50S ribosomal protein L22
+Macromolecule #25: 50S ribosomal protein L23
+Macromolecule #26: 50S ribosomal protein L24
+Macromolecule #27: 50S ribosomal protein L25
+Macromolecule #28: 50S ribosomal protein L27
+Macromolecule #29: 50S ribosomal protein L28
+Macromolecule #30: 50S ribosomal protein L29
+Macromolecule #31: 50S ribosomal protein L30
+Macromolecule #32: 50S ribosomal protein L32
+Macromolecule #7: 23S ribosomal RNA
+Macromolecule #8: 5S ribosomal RNA
+Macromolecule #33: ZINC ION
+Macromolecule #34: 2-acetamido-2-deoxy-alpha-D-galactopyranose
+Macromolecule #35: MAGNESIUM ION
+Macromolecule #36: SPERMINE
+Macromolecule #37: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R3/3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 3 | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | 8 OD260/mL |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.9 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)