+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of sweet potato feathery mottle virus VLP | |||||||||||||||
 Map data Map data | Sharpened map. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | plant virus / coat protein / VIRUS LIKE PARTICLE | |||||||||||||||
| Function / homology | Potyvirus coat protein / Potyvirus coat protein / viral capsid / Genome polyprotein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Sweet potato feathery mottle virus / Sweet potato feathery mottle virus /  Ipomoea batatas (sweet potato) Ipomoea batatas (sweet potato) | |||||||||||||||
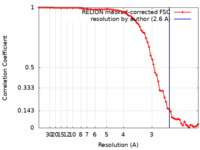
| Method | helical reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||||||||
 Authors Authors | Javed A / Byrne JM / Ranson N / Lomonosoff G | |||||||||||||||
| Funding support |  Spain, Spain,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: CryoEM and stability analysis of virus-like particles of potyvirus and ipomovirus infecting a common host. Authors: Ornela Chase / Abid Javed / Matthew J Byrne / Eva C Thuenemann / George P Lomonossoff / Neil A Ranson / Juan José López-Moya /   Abstract: Sweet potato feathery mottle virus (SPFMV) and Sweet potato mild mottle virus (SPMMV) are members of the genera Potyvirus and Ipomovirus, family Potyviridae, sharing Ipomoea batatas as common host, ...Sweet potato feathery mottle virus (SPFMV) and Sweet potato mild mottle virus (SPMMV) are members of the genera Potyvirus and Ipomovirus, family Potyviridae, sharing Ipomoea batatas as common host, but transmitted, respectively, by aphids and whiteflies. Virions of family members consist of flexuous rods with multiple copies of a single coat protein (CP) surrounding the RNA genome. Here we report the generation of virus-like particles (VLPs) by transient expression of the CPs of SPFMV and SPMMV in the presence of a replicating RNA in Nicotiana benthamiana. Analysis of the purified VLPs by cryo-electron microscopy, gave structures with resolutions of 2.6 and 3.0 Å, respectively, showing a similar left-handed helical arrangement of 8.8 CP subunits per turn with the C-terminus at the inner surface and a binding pocket for the encapsidated ssRNA. Despite their similar architecture, thermal stability studies reveal that SPMMV VLPs are more stable than those of SPFMV. | |||||||||||||||
| History |
|
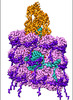
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15345.map.gz emd_15345.map.gz | 11.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15345-v30.xml emd-15345-v30.xml emd-15345.xml emd-15345.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15345_fsc.xml emd_15345_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15345.png emd_15345.png | 140.3 KB | ||
| Filedesc metadata |  emd-15345.cif.gz emd-15345.cif.gz | 6.3 KB | ||
| Others |  emd_15345_half_map_1.map.gz emd_15345_half_map_1.map.gz emd_15345_half_map_2.map.gz emd_15345_half_map_2.map.gz | 26.2 MB 26.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15345 http://ftp.pdbj.org/pub/emdb/structures/EMD-15345 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15345 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15345 | HTTPS FTP |
-Related structure data
| Related structure data |  8acbMC  8accC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15345.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15345.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||
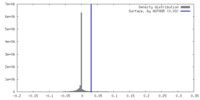
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half 1 map.
| File | emd_15345_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half 1 map. | ||||||||||||
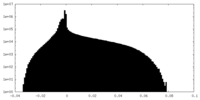
| Projections & Slices |
| ||||||||||||
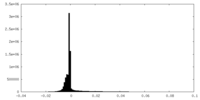
| Density Histograms |
-Half map: Half 2 map
| File | emd_15345_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half 2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
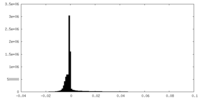
| Density Histograms |
- Sample components
Sample components
-Entire : Sweet potato feathery mottle virus
| Entire | Name:  Sweet potato feathery mottle virus Sweet potato feathery mottle virus |
|---|---|
| Components |
|
-Supramolecule #1: Sweet potato feathery mottle virus
| Supramolecule | Name: Sweet potato feathery mottle virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 12844 / Sci species name: Sweet potato feathery mottle virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Ipomoea batatas (sweet potato) Ipomoea batatas (sweet potato) |
| Molecular weight | Theoretical: 380 KDa |
| Virus shell | Shell ID: 1 / Diameter: 130.0 Å |
-Macromolecule #1: Genome polyprotein
| Macromolecule | Name: Genome polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sweet potato feathery mottle virus Sweet potato feathery mottle virus |
| Molecular weight | Theoretical: 25.209426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VPRVKMNANK KRQPMVNGRA IINFQHLSTY EPEQFEVANT RSTQEQFQAW YEGVKGDYGV DDTGMGILLN GLMVWCIENG TSPNINGVW TMMDGDEQVT YPIKPLLDHA VPTFRQIMTH FSDVAEAYIE MRNRTKAYMP RYGLQRNLTD MSLARYAFDF Y ELHSTTPA ...String: VPRVKMNANK KRQPMVNGRA IINFQHLSTY EPEQFEVANT RSTQEQFQAW YEGVKGDYGV DDTGMGILLN GLMVWCIENG TSPNINGVW TMMDGDEQVT YPIKPLLDHA VPTFRQIMTH FSDVAEAYIE MRNRTKAYMP RYGLQRNLTD MSLARYAFDF Y ELHSTTPA RAKEAHLQMK AAALKNARNR LFGLDGNVST QEEDTERHTT TDVTRNIHNL LG UniProtKB: Genome polyprotein |
-Macromolecule #2: Single-stranded RNA
| Macromolecule | Name: Single-stranded RNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Ipomoea batatas (sweet potato) Ipomoea batatas (sweet potato) |
| Molecular weight | Theoretical: 1.485872 KDa |
| Sequence | String: UUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Concentration: 50.0 mM / Component - Formula: NaPO4 / Component - Name: Sodium Phosphate |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3276 / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 140.9 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-8acb: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)