+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | ||||||||||||
 マップデータ マップデータ | Postprocessed map from Relion | ||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | ICOSAHEDRAL VIRUS / AUTO-CATALYTIC CLEAVAGE / VIRUS MATURATION / VIRUS-LIKE PARTICLE / VIRUS LIKE PARTICLE | ||||||||||||
| 機能・相同性 | Peptidase N2 / Peptidase family A21 / virion component / Viral coat protein subunit / p70 機能・相同性情報 機能・相同性情報 | ||||||||||||
| 生物種 |   Nudaurelia capensis omega virus (ウイルス) Nudaurelia capensis omega virus (ウイルス) | ||||||||||||
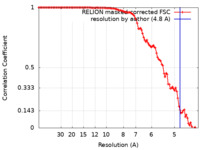
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.8 Å | ||||||||||||
 データ登録者 データ登録者 | Castells-Graells R / Hesketh EL / Johnson JE / Ranson NA / Lawson DM / Lomonossoff GP | ||||||||||||
| 資金援助 |  英国, 3件 英国, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: To be published ジャーナル: To be publishedタイトル: Decoding virus maturation with cryo-EM structures of intermediates 著者: Castells-Graells R / Hesketh EL / Johnson JE / Ranson NA / Lawson DM / Lomonossoff GP | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_15209.map.gz emd_15209.map.gz | 93.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-15209-v30.xml emd-15209-v30.xml emd-15209.xml emd-15209.xml | 17.7 KB 17.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_15209_fsc.xml emd_15209_fsc.xml | 10.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_15209.png emd_15209.png | 193.4 KB | ||
| マスクデータ |  emd_15209_msk_1.map emd_15209_msk_1.map | 103 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-15209.cif.gz emd-15209.cif.gz | 6.2 KB | ||
| その他 |  emd_15209_half_map_1.map.gz emd_15209_half_map_1.map.gz emd_15209_half_map_2.map.gz emd_15209_half_map_2.map.gz | 79.8 MB 80 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15209 http://ftp.pdbj.org/pub/emdb/structures/EMD-15209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15209 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_15209.map.gz / 形式: CCP4 / 大きさ: 103 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_15209.map.gz / 形式: CCP4 / 大きさ: 103 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Postprocessed map from Relion | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 2.13 Å | ||||||||||||||||||||||||||||||||||||
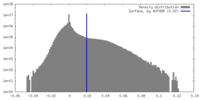
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_15209_msk_1.map emd_15209_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Refine3D half map 1 from Relion
| ファイル | emd_15209_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Refine3D half map 1 from Relion | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Refine3D half map 2 from Relion
| ファイル | emd_15209_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Refine3D half map 2 from Relion | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Nudaurelia capensis omega virus
| 全体 | 名称:   Nudaurelia capensis omega virus (ウイルス) Nudaurelia capensis omega virus (ウイルス) |
|---|---|
| 要素 |
|
-超分子 #1: Nudaurelia capensis omega virus
| 超分子 | 名称: Nudaurelia capensis omega virus / タイプ: virus / ID: 1 / 親要素: 0 / 含まれる分子: all / 詳細: expressed in Spodoptera frugiperda Sf21 cells / NCBI-ID: 12541 / 生物種: Nudaurelia capensis omega virus / ウイルスタイプ: VIRUS-LIKE PARTICLE / ウイルス・単離状態: OTHER / ウイルス・エンベロープ: No / ウイルス・中空状態: No |
|---|---|
| 宿主 | 生物種:  Gonimbrasia cytherea (蝶・蛾) Gonimbrasia cytherea (蝶・蛾) |
| 分子量 | 理論値: 16.76 MDa |
| ウイルス殻 | Shell ID: 1 / 名称: coat / 直径: 460.0 Å / T番号(三角分割数): 4 |
-分子 #1: p70
| 分子 | 名称: p70 / タイプ: protein_or_peptide / ID: 1 / コピー数: 4 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:   Nudaurelia capensis omega virus (ウイルス) Nudaurelia capensis omega virus (ウイルス) |
| 分子量 | 理論値: 69.891953 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MDSNSASGKR RSRNVRIAAN TVNVAPKQRQ ARGRRARSRA NNIDNVTAAA QELGQSLDAN VITFPTNVAT MPEFRSWARG KLDIDQDSI GWYFKYLDPA GATESARAVG EYSKIPDGLV KFSVDAEIRE IYNEECPTVS DASIPLDGAQ WSLSIISYPM F RTAYFAVA ...文字列: MDSNSASGKR RSRNVRIAAN TVNVAPKQRQ ARGRRARSRA NNIDNVTAAA QELGQSLDAN VITFPTNVAT MPEFRSWARG KLDIDQDSI GWYFKYLDPA GATESARAVG EYSKIPDGLV KFSVDAEIRE IYNEECPTVS DASIPLDGAQ WSLSIISYPM F RTAYFAVA NVDNKEISLD VTNDLIVWLN NLASWRDVVD SGQWFTFSDD PTWFVRIRVL HPTYDLPDPT EGLLRTVSDY RL TYKSITC EANMPTLVDQ GFWIGGHYAL TPIATTQNAV EGSGFVHPFN VTRPGIAAGV TLTWASMPPG GSAPSGDPAW IPD STTQFQ WRHGGFDAPT GVITYTIPRG YTMQYFDTTT NEWNGFANPD DVVTFGQTGG AAGTNATITI TAPTVTLTIL ATTT SAANV INFRNLDAET TAASNRSEVP LPPLTFGQTA PNNPKIEQTL VKDTLGSYLV HSKMRNPVFQ LTPASSFGAI SFTNP GFDR NLDLPGFGGI RDSLDVNMST AVCHFRSLSK SCSIVTKTYQ GWEGVTNVNT PFGQFAHSGL LKNDEILCLA DDLATR LTG VYGATDNFAA AVLAFAANML TSVLKSEATT SVIKELGNQA TGLANQGLAR LPGLLASIPG KIAARVRARR DRRRAAR MN NN UniProtKB: p70 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.3 mg/mL |
|---|---|
| 緩衝液 | pH: 6.25 |
| グリッド | 材質: COPPER / メッシュ: 400 |
| 凍結 | 凍結剤: ETHANE / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON III (4k x 4k) 検出モード: INTEGRATING / 撮影したグリッド数: 1 / 実像数: 10528 / 平均露光時間: 1.0 sec. / 平均電子線量: 50.4 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.2 µm / 最小 デフォーカス(公称値): 0.8 µm / 倍率(公称値): 75000 |
| 試料ステージ | ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)