[English] 日本語
 Yorodumi
Yorodumi- EMDB-14771: Amyloid fibril from human systemic AA amyloidosis (vascular variant) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Amyloid fibril from human systemic AA amyloidosis (vascular variant) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloids / Vascular AA / SAA / Cryo-EM / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationlymphocyte chemotaxis / Scavenging by Class B Receptors / positive regulation of interleukin-1 production / high-density lipoprotein particle / Formyl peptide receptors bind formyl peptides and many other ligands / regulation of protein secretion / macrophage chemotaxis / TRAF6 mediated NF-kB activation / Advanced glycosylation endproduct receptor signaling / cytoplasmic microtubule ...lymphocyte chemotaxis / Scavenging by Class B Receptors / positive regulation of interleukin-1 production / high-density lipoprotein particle / Formyl peptide receptors bind formyl peptides and many other ligands / regulation of protein secretion / macrophage chemotaxis / TRAF6 mediated NF-kB activation / Advanced glycosylation endproduct receptor signaling / cytoplasmic microtubule / neutrophil chemotaxis / endocytic vesicle lumen / positive regulation of cell adhesion / positive regulation of cytokine production / acute-phase response / TAK1-dependent IKK and NF-kappa-B activation / G protein-coupled receptor binding / platelet activation / negative regulation of inflammatory response / heparin binding / positive regulation of cytosolic calcium ion concentration / Interleukin-4 and Interleukin-13 signaling / G alpha (i) signalling events / G alpha (q) signalling events / Amyloid fiber formation / extracellular exosome / extracellular region Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
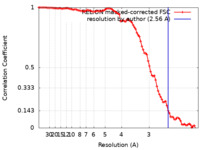
| Method | helical reconstruction / cryo EM / Resolution: 2.56 Å | |||||||||
 Authors Authors | Banerjee S / Schmidt M / Faendrich M | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Amyloid fibril structure from the vascular variant of systemic AA amyloidosis. Authors: Sambhasan Banerjee / Julian Baur / Christoph Daniel / Peter Benedikt Pfeiffer / Manuel Hitzenberger / Lukas Kuhn / Sebastian Wiese / Johan Bijzet / Christian Haupt / Kerstin U Amann / Martin ...Authors: Sambhasan Banerjee / Julian Baur / Christoph Daniel / Peter Benedikt Pfeiffer / Manuel Hitzenberger / Lukas Kuhn / Sebastian Wiese / Johan Bijzet / Christian Haupt / Kerstin U Amann / Martin Zacharias / Bouke P C Hazenberg / Gunilla T Westermark / Matthias Schmidt / Marcus Fändrich /    Abstract: Systemic AA amyloidosis is a debilitating protein misfolding disease in humans and animals. In humans, it occurs in two variants that are called 'vascular' and 'glomerular', depending on the main ...Systemic AA amyloidosis is a debilitating protein misfolding disease in humans and animals. In humans, it occurs in two variants that are called 'vascular' and 'glomerular', depending on the main amyloid deposition site in the kidneys. Using cryo electron microscopy, we here show the amyloid fibril structure underlying the vascular disease variant. Fibrils purified from the tissue of such patients are mainly left-hand twisted and contain two non-equal stacks of fibril proteins. They contrast in these properties to the fibrils from the glomerular disease variant which are right-hand twisted and consist of two structurally equal stacks of fibril proteins. Our data demonstrate that the different disease variants in systemic AA amyloidosis are associated with different fibril morphologies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14771.map.gz emd_14771.map.gz | 8.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14771-v30.xml emd-14771-v30.xml emd-14771.xml emd-14771.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14771_fsc.xml emd_14771_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14771.png emd_14771.png | 21.8 KB | ||
| Filedesc metadata |  emd-14771.cif.gz emd-14771.cif.gz | 4.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14771 http://ftp.pdbj.org/pub/emdb/structures/EMD-14771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14771 | HTTPS FTP |
-Related structure data
| Related structure data |  7zkyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14771.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14771.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Amyloid fibril from human systemic AA amyloidosis (vascular variant)
| Entire | Name: Amyloid fibril from human systemic AA amyloidosis (vascular variant) |
|---|---|
| Components |
|
-Supramolecule #1: Amyloid fibril from human systemic AA amyloidosis (vascular variant)
| Supramolecule | Name: Amyloid fibril from human systemic AA amyloidosis (vascular variant) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Amyloid protein A
| Macromolecule | Name: Amyloid protein A / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.775502 KDa |
| Sequence | String: SFFSFLGEAF DGARDMWRAY SDMREANYIG SDKYFHARGN YDAAKRGPGG VWAAEAISDA RENIQRFF UniProtKB: Serum amyloid A-1 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 10.0 sec. / Average electron dose: 42.84 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7zky: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)