+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of hnRNPDL amyloid fibrils | ||||||||||||
 Map data Map data | sharped | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Amyloid / Protein aggregation / Alternative splicing / Exon / Prion-like / LGMD1G / cryoEM structure / RNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpoly(G) binding / poly(A) binding / RNA processing / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / spliceosomal complex / single-stranded DNA binding / double-stranded DNA binding / regulation of gene expression / chromatin / DNA binding ...poly(G) binding / poly(A) binding / RNA processing / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / spliceosomal complex / single-stranded DNA binding / double-stranded DNA binding / regulation of gene expression / chromatin / DNA binding / RNA binding / nucleoplasm / nucleus / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
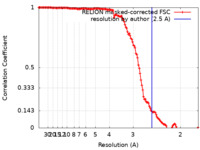
| Method | helical reconstruction / cryo EM / Resolution: 2.5 Å | ||||||||||||
 Authors Authors | Chaves-Sanjuan A / Garcia-Pardo J / Bartolome-Nafria A / Gil-Garcia M / Visentin C / Bolognesi M / Ricagno S / Ventura S | ||||||||||||
| Funding support |  Spain, 3 items Spain, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structure of hnRNPDL-2 fibrils, a functional amyloid associated with limb-girdle muscular dystrophy D3. Authors: Javier Garcia-Pardo / Andrea Bartolomé-Nafría / Antonio Chaves-Sanjuan / Marcos Gil-Garcia / Cristina Visentin / Martino Bolognesi / Stefano Ricagno / Salvador Ventura /   Abstract: hnRNPDL is a ribonucleoprotein (RNP) involved in transcription and RNA-processing that hosts missense mutations causing limb-girdle muscular dystrophy D3 (LGMD D3). Mammalian-specific alternative ...hnRNPDL is a ribonucleoprotein (RNP) involved in transcription and RNA-processing that hosts missense mutations causing limb-girdle muscular dystrophy D3 (LGMD D3). Mammalian-specific alternative splicing (AS) renders three natural isoforms, hnRNPDL-2 being predominant in humans. We present the cryo-electron microscopy structure of full-length hnRNPDL-2 amyloid fibrils, which are stable, non-toxic, and bind nucleic acids. The high-resolution amyloid core consists of a single Gly/Tyr-rich and highly hydrophilic filament containing internal water channels. The RNA binding domains are located as a solenoidal coat around the core. The architecture and activity of hnRNPDL-2 fibrils are reminiscent of functional amyloids, our results suggesting that LGMD D3 might be a loss-of-function disease associated with impaired fibrillation. Strikingly, the fibril core matches exon 6, absent in the soluble hnRNPDL-3 isoform. This provides structural evidence for AS controlling hnRNPDL assembly by precisely including/skipping an amyloid exon, a mechanism that holds the potential to generate functional diversity in RNPs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14738.map.gz emd_14738.map.gz | 9.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14738-v30.xml emd-14738-v30.xml emd-14738.xml emd-14738.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14738_fsc.xml emd_14738_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_14738.png emd_14738.png | 68.8 KB | ||
| Filedesc metadata |  emd-14738.cif.gz emd-14738.cif.gz | 6.1 KB | ||
| Others |  emd_14738_additional_1.map.gz emd_14738_additional_1.map.gz emd_14738_half_map_1.map.gz emd_14738_half_map_1.map.gz emd_14738_half_map_2.map.gz emd_14738_half_map_2.map.gz | 163 MB 164.4 MB 164.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14738 http://ftp.pdbj.org/pub/emdb/structures/EMD-14738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14738 | HTTPS FTP |
-Validation report
| Summary document |  emd_14738_validation.pdf.gz emd_14738_validation.pdf.gz | 617.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14738_full_validation.pdf.gz emd_14738_full_validation.pdf.gz | 616.8 KB | Display | |
| Data in XML |  emd_14738_validation.xml.gz emd_14738_validation.xml.gz | 21.1 KB | Display | |
| Data in CIF |  emd_14738_validation.cif.gz emd_14738_validation.cif.gz | 27.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14738 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14738 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14738 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14738 | HTTPS FTP |
-Related structure data
| Related structure data |  7zirMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14738.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14738.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharped | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
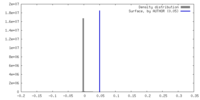
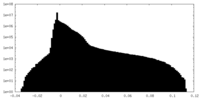
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharped
| File | emd_14738_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharped | ||||||||||||
| Projections & Slices |
| ||||||||||||
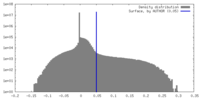
| Density Histograms |
-Half map: half 1
| File | emd_14738_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
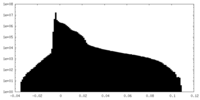
| Density Histograms |
-Half map: half 2
| File | emd_14738_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
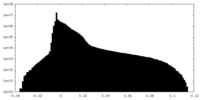
| Density Histograms |
- Sample components
Sample components
-Entire : hnRNPDL-2 amyloid fibrils
| Entire | Name: hnRNPDL-2 amyloid fibrils |
|---|---|
| Components |
|
-Supramolecule #1: hnRNPDL-2 amyloid fibrils
| Supramolecule | Name: hnRNPDL-2 amyloid fibrils / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Fibril structure of the recombinantly produced full-lenght Heterogeneous ribonucleoprotein D-like (hnRNPDL) isoform 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Heterogeneous nuclear ribonucleoprotein D-like
| Macromolecule | Name: Heterogeneous nuclear ribonucleoprotein D-like / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.804266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSL QDSEVNQEAK PEVKPEVKPE THINLKVSDG SSEIFFKIKK TTPLRRLMEA FAKRQGKEMD SLTFLYDGIE IQADQTPED LDMEDNDIIE AHREQIGGED MNEYSNIEEF AEGSKINASK NQQDDGKMFI GGLSWDTSKK DLTEYLSRFG E VVDCTIKT ...String: MHHHHHHGSL QDSEVNQEAK PEVKPEVKPE THINLKVSDG SSEIFFKIKK TTPLRRLMEA FAKRQGKEMD SLTFLYDGIE IQADQTPED LDMEDNDIIE AHREQIGGED MNEYSNIEEF AEGSKINASK NQQDDGKMFI GGLSWDTSKK DLTEYLSRFG E VVDCTIKT DPVTGRSRGF GFVLFKDAAS VDKVLELKEH KLDGKLIDPK RAKALKGKEP PKKVFVGGLS PDTSEEQIKE YF GAFGEIE NIELPMDTKT NERRGFCFIT YTDEEPVKKL LESRYHQIGS GKCEIKVAQP KEVYRQQQQQ QKGGRGAAAG GRG GTRGRG RGQGQNWNQG FNNYYDQGYG NYNSAYGGDQ NYSGYGGYDY TGYNYGNYGY GQGYADYSGQ QSTYGKASRG GGNH QNNYQ PY UniProtKB: Heterogeneous nuclear ribonucleoprotein D-like |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 50 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.23 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 / Component:
| ||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV | ||||||
| Details | Diluted sample of hnRNPDL2 amyloid fibrils formed under native conditions. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number grids imaged: 1 / Number real images: 1114 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 120000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
|---|---|
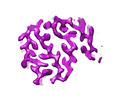
| Output model |  PDB-7zir: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)