[English] 日本語
 Yorodumi
Yorodumi- EMDB-14407: Single particle structure of keyhole limpet hemocyanin obtained v... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle structure of keyhole limpet hemocyanin obtained via iDPC scanning transmission electron microscopy | |||||||||
 Map data Map data | sharpened map from cryosparc local refinement v3.3.1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | iDPC / STEM / KLH / single particle analysis / OXYGEN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationoxygen carrier activity / oxidoreductase activity / extracellular region / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Megathura crenulata (invertebrata) Megathura crenulata (invertebrata) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.51 Å | |||||||||
 Authors Authors | Mann D / Lazic I / Wirix M / de Haas F / Sachse C | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2022 Journal: Nat Methods / Year: 2022Title: Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution. Authors: Ivan Lazić / Maarten Wirix / Max Leo Leidl / Felix de Haas / Daniel Mann / Maximilian Beckers / Evgeniya V Pechnikova / Knut Müller-Caspary / Ricardo Egoavil / Eric G T Bosch / Carsten Sachse /   Abstract: In electron cryomicroscopy (cryo-EM), molecular images of vitrified biological samples are obtained by conventional transmission microscopy (CTEM) using large underfocuses and subsequently ...In electron cryomicroscopy (cryo-EM), molecular images of vitrified biological samples are obtained by conventional transmission microscopy (CTEM) using large underfocuses and subsequently computationally combined into a high-resolution three-dimensional structure. Here, we apply scanning transmission electron microscopy (STEM) using the integrated differential phase contrast mode also known as iDPC-STEM to two cryo-EM test specimens, keyhole limpet hemocyanin (KLH) and tobacco mosaic virus (TMV). The micrographs show complete contrast transfer to high resolution and enable the cryo-EM structure determination for KLH at 6.5 Å resolution, as well as for TMV at 3.5 Å resolution using single-particle reconstruction methods, which share identical features with maps obtained by CTEM of a previously acquired same-sized TMV data set. These data show that STEM imaging in general, and in particular the iDPC-STEM approach, can be applied to vitrified single-particle specimens to determine near-atomic resolution cryo-EM structures of biological macromolecules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14407.map.gz emd_14407.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14407-v30.xml emd-14407-v30.xml emd-14407.xml emd-14407.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
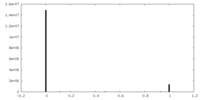
| FSC (resolution estimation) |  emd_14407_fsc.xml emd_14407_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_14407.png emd_14407.png | 259.7 KB | ||
| Masks |  emd_14407_msk_1.map emd_14407_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14407.cif.gz emd-14407.cif.gz | 7 KB | ||
| Others |  emd_14407_half_map_1.map.gz emd_14407_half_map_1.map.gz emd_14407_half_map_2.map.gz emd_14407_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14407 http://ftp.pdbj.org/pub/emdb/structures/EMD-14407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14407 | HTTPS FTP |
-Related structure data
| Related structure data |  7q22C  7q23C  7q2qC  7q2rC  7q2sC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14407.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14407.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map from cryosparc local refinement v3.3.1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.44 Å | ||||||||||||||||||||||||||||||||||||
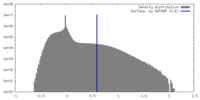
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14407_msk_1.map emd_14407_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_14407_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_14407_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : keyhole limpet hemocyanin
| Entire | Name: keyhole limpet hemocyanin |
|---|---|
| Components |
|
-Supramolecule #1: keyhole limpet hemocyanin
| Supramolecule | Name: keyhole limpet hemocyanin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Megathura crenulata (invertebrata) Megathura crenulata (invertebrata) |
-Macromolecule #1: keyhole limpet hemocyanin
| Macromolecule | Name: keyhole limpet hemocyanin / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Megathura crenulata (invertebrata) Megathura crenulata (invertebrata) |
| Sequence | String: MLSVRLLIVV LALANAENLV RKSVEHLTQE ETLDLQAALR ELQMDSSSIG FQKIAAAHGA PASCVHKDTS IACCIHGMPT FPHWHRAYVV HMERALQTKR RTSGLPYWDW TEPITQLPSL AADPVYIDSQ GGKAHTNYWY RGNIDFLDKK TNRAVDDRLF EKVKPGQHTH ...String: MLSVRLLIVV LALANAENLV RKSVEHLTQE ETLDLQAALR ELQMDSSSIG FQKIAAAHGA PASCVHKDTS IACCIHGMPT FPHWHRAYVV HMERALQTKR RTSGLPYWDW TEPITQLPSL AADPVYIDSQ GGKAHTNYWY RGNIDFLDKK TNRAVDDRLF EKVKPGQHTH LMESVLDALE QDEFCKFEIQ FELAHNAIHY LVGGKHDYSM ANLEYTAYDP IFFLHHSNVD RIFAIWQRLQ ELRNKDPKAM DCAQELLHQK MEPFSWEDND IPLTNEHSTP ADLFDYCELH YDYDTLNLNG MTPEELKTYL DERSSRARAF ASFRLKGFGG SANVFVYVCI PDDNDRNDDH CEKAGDFFVL GGPSEMKWQF YRPYLFDLSD TVHKMGMKLD GHYTVKAELF SVNGTALPDD LLPHPVVVHH PEKGFTDPPV KHHQSANLLV RKNINDLTRE EVLNLREAFH KFQEDRSVDG YQATAEYHGL PARCPRPDAK DRYACCVHGM PIFPHWHRLF VTQVEDALVG RGATIGIPYW DWTEPMTHIP GLAGNKTYVD SHGASHTNPF HSSVIAFEEN APHTKRQIDQ RLFKPATFGH HTDLFNQILY AFEQEDYCDF EVQFEITHNT IHAWTGGSEH FSMSSLHYTA FDPLFYFHHS NVDRLWAVWQ ALQMRRHKPY RAHCAISLEH MHLKPFAFSS PLNNNEKTHA NAMPNKIYDY ENVLHYTYED LTFGGISLEN IEKMIHENQQ EDRIYAGFLL AGIRTSANVD IFIKTTDSVQ HKAGTFAVLG GSKEMKWGFD RVFKFDITHV LKDLDLTADG DFEVTVDITE VDGTKLASSL IPHASVIREH ARVKFDKVPR SRLIRKNVDR LSPEEMNELR KALALLKEDK SAGGFQQLGA FHGEPKWCPS PEASKKFACC VHGMSVFPHW HRLLTVQSEN ALRRHGYDGA LPYWDWTSPL NHLPELADHE KYVDPEDGVE KHNPWFDGHI DTVDKTTTRS VQNKLFEQPE FGHYTSIAKQ VLLALEQDNF CDFEIQYEIA HNYIHALVGG AQPYGMASLR YTAFDPLFYL HHSNTDRIWA IWQALQKYRG KPYNVANCAV TSMREPLQPF GLSANINTDH VTKEHSVPFN VFDYKTNFNY EYDTLEFNGL SISQLNKKLE AIKSQDRFFA GFLLSGFKKS SLVKFNICTD SSNCHPAGEF YLLGDENEMP WAYDRVFKYD ITEKLHDLKL HAEDHFYIDY EVFDLKPASL GKDLFKQPSV IHEPRIGHHE GEVYQAEVTS ANRIRKNIEN LSLGELESLR AAFLEIENDG TYESIAKFHG SPGLCQLNGN PISCCVHGMP TFPHWHRLYV VVVENALLKK GSSVAVPYWD WTKRIEHLPH LISDATYYNS RQHHYETNPF HHGKITHENE ITTRDPKDSL FHSDYFYEQV LYALEQDNFC DFEIQLEILH NALHSLLGGK GKYSMSNLDY AAFDPVFFLH HATTDRIWAI WQDLQRFRKR PYREANCAIQ LMHTPLQPFD KSDNNDEATK THATPHDGFE YQNSFGYAYD NLELNHYSIP QLDHMLQERK RHDRVFAGFL LHNIGTSADG HVFVCLPTGE HTKDCSHEAG MFSILGGQTE MSFVFDRLYK LDITKALKKN GVHLQGDFDL EIEITAVNGS HLDSHVIHSP TILFEAGTDS AHTDDGHTEP VMIRKDITQL DKRQQLSLVK ALESMKADHS SDGFQAIASF HALPPLCPSP AASKRFACCV HGMATFPQWH RLYTVQFQDS LRKHGAVVGL PYWDWTLPRS ELPELLTVST IHDPETGRDI PNPFIGSKIE FEGENVHTKR DINRDRLFQG STKTHHNWFI EQALLALEQT NYCDFEVQFE IMHNGVHTWV GGKEPYGIGH LHYASYDPLF YIHHSQTDRI WAIWQSLQRF RGLSGSEANC AVNLMKTPLK PFSFGAPYNL NDHTHDFSKP EDTFDYQKFG YIYDTLEFAG WSIRGIDHIV RNRQEHSRVF AGFLLEGFGT SATVDFQVCR TAGDCEDAGY FTVLGGEKEM PWAFDRLYKY DITETLDKMN LRHDEIFQIE VTITSYDGTV LDSGLIPTPS IIYDPAHHDI SSHHLSLNKV RHDLSTLSER DIGSLKYALS SLQADTSADG FAAIASFHGL PAKCNDSHNN EVACCIHGMP TFPHWHRLYT LQFEQALRRH GSSVAVPYWD WTKPIHNIPH LFTDKEYYDV WRNKVMPNPF ARGYVPSHDT YTVRDVQEGL FHLTSTGEHS ALLNQALLAL EQHDYCDFAV QFEVMHNTIH YLVGGPQVYS LSSLHYASYD PIFFIHHSFV DKVWAVWQAL QEKRGLPSDR ADCAVSLMTQ NMRPFHYEIN HNQFTKKHAV PNDVFKYELL GYRYDNLEIG GMNLHEIEKE IKDKQHHVRV FAGFLLHGIR TSADVQFQIC KTSEDCHHGG QIFVLGGTKE MAWAYNRLFK YDITHALHDA HITPEDVFHP SEPFFIKVSV TAVNGTVLPA SILHAPTIIY EPGLDHHEDH HSSSMAGHGV RKEINTLTTA EVDNLKDAMR AVMADHGPNG YQAIAAFHGN PPMCPMPDGK NYSCCTHGMA TFPHWHRLYT KQMEDALTAH GARVGLPYWD GTTAFTALPT FVTDEEDNPF HHGHIDYLGV DTTRSPRDKL FNDPERGSES FFYRQVLLAL EQTDFCQFEV QFEITHNAIH SWTGGLTPYG MSTLEYTTYD PLFWLHHANT DRIWAIWQAL QEYRGLPYDH ANCEIQAMKR PLRPFSDPIN HNAFTHSNAK PTDVFEYSRF NFQYDNLRFH GMTIKKLEHE LEKQKEEDRT FAAFLLHGIK KSADVSFDVC NHDGECHFAG TFAILGGEHE MPWSFDRLFR YDITQVLKQM HLEYDSDFTF HMRIIDTSGK QLPSDLIKMP TVEHSPGGKH HEKHHEDHHE DILVRKNIHS LSHHEAEELR DALYKLQNDE SHGGYEHIAG FHGYPNLCPE KGDEKYPCCV HGMSIFPHWH RLHTIQFERA LKKHGSHLGI PYWDWTQTIS SLPTFFADSG NNNPFFKYHI RSINQDTVRD VNEAIFQQTK FGEFSSIFYL ALQALEEDNY CDFEVQYEIL HNEVHALIGG AEKYSMSTLE YSAFDPYFMI HHASLDKIWI IWQELQKRRV KPAHAGSCAG DIMHVPLHPF NYESVNNDDF TRENSLPNAV VDSHRFNYKY DNLNLHGHNI EELEEVLRSL RLKSRVFAGF VLSGIRTTAV VKVYIKSGTD SDDEYAGSFV ILGGAKEMPW AYERLYRFDI TETVHNLNLT DDHVKFRFDL KKYDHTELDA SVLPAPIIVR RPNNAVFDII EIPIGKDVNL PPKVVVKRGT KIMFMSVDEA VTTPMLNLGS YTAMFKCKVP PFSFHAFELG KMYSVESGDY FMTASTTELC NDNNLRIHVH VDDE UniProtKB: Hemocyanin 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 35.0 e/Å2 Details: STEM imaging was conducted using a Thermo Fisher Scientific Titan Krios G4, operated at 300 kV. The column was equipped with a standard high-brightness field emission gun (XFEG), three- ...Details: STEM imaging was conducted using a Thermo Fisher Scientific Titan Krios G4, operated at 300 kV. The column was equipped with a standard high-brightness field emission gun (XFEG), three-condenser lens system, C-TWIN objective lens with wide-gap pole piece (11 mm and Cs = 2.7mm), Panther segmented STEM detector, a Ceta camera and Falcon 4 direct electron detection (DED) camera. A combination of different C2 apertures (20 mum and 50 mum) and C2/C3 lens current ratios were used to create different CSA of the beam. Alignment for STEM was done by aligning beam shift, beam tilt pivot points and beam tilt in STEM mode for the different convergence angles. For accurate determination of the COM, a de-scan alignment has additionally been performed. The CSA of the applied beams were measured with high precision using the Au cross-grating and Ceta camera by first recording the radii of the gold diffraction rings for calibration and subsequently measuring the radius of the bright-field (BF) disc. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: OTHER / Nominal defocus max: 0.0 µm / Nominal defocus min: 0.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)