+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Amyloid fibril from the antimicrobial peptide aurein 3.3 | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | antimicrobial peptide / amyloid / filament / cross-beta / PROTEIN FIBRIL | ||||||||||||||||||
| Function / homology | Aurein antibiotic peptide family / Aurein-like antibiotic peptide / defense response to bacterium / extracellular region / Aurein-3.3 Function and homology information Function and homology information | ||||||||||||||||||
| Biological species | Ranoidea raniformis (blue-thighed treefrog) | ||||||||||||||||||
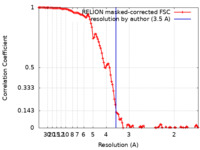
| Method | helical reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||||||||
 Authors Authors | Buecker R / Seuring C | ||||||||||||||||||
| Funding support | 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: The Cryo-EM structures of two amphibian antimicrobial cross-β amyloid fibrils. Authors: Robert Bücker / Carolin Seuring / Cornelia Cazey / Katharina Veith / Maria García-Alai / Kay Grünewald / Meytal Landau /    Abstract: The amyloid-antimicrobial link hypothesis is based on antimicrobial properties found in human amyloids involved in neurodegenerative and systemic diseases, along with amyloidal structural properties ...The amyloid-antimicrobial link hypothesis is based on antimicrobial properties found in human amyloids involved in neurodegenerative and systemic diseases, along with amyloidal structural properties found in antimicrobial peptides (AMPs). Supporting this hypothesis, we here determined the fibril structure of two AMPs from amphibians, uperin 3.5 and aurein 3.3, by cryogenic electron microscopy (cryo-EM), revealing amyloid cross-β fibrils of mated β-sheets at atomic resolution. Uperin 3.5 formed a 3-blade symmetrical propeller of nine peptides per fibril layer including tight β-sheet interfaces. This cross-β cryo-EM structure complements the cross-α fibril conformation previously determined by crystallography, substantiating a secondary structure switch mechanism of uperin 3.5. The aurein 3.3 arrangement consisted of six peptides per fibril layer, all showing kinked β-sheets allowing a rounded compactness of the fibril. The kinked β-sheets are similar to LARKS (Low-complexity, Amyloid-like, Reversible, Kinked Segments) found in human functional amyloids. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14168.map.gz emd_14168.map.gz | 125 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14168-v30.xml emd-14168-v30.xml emd-14168.xml emd-14168.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14168_fsc.xml emd_14168_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14168.png emd_14168.png | 123.9 KB | ||
| Masks |  emd_14168_msk_1.map emd_14168_msk_1.map | 134.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14168.cif.gz emd-14168.cif.gz | 5.7 KB | ||
| Others |  emd_14168_additional_1.map.gz emd_14168_additional_1.map.gz emd_14168_half_map_1.map.gz emd_14168_half_map_1.map.gz emd_14168_half_map_2.map.gz emd_14168_half_map_2.map.gz | 70.6 MB 105.8 MB 106 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14168 http://ftp.pdbj.org/pub/emdb/structures/EMD-14168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14168 | HTTPS FTP |
-Related structure data
| Related structure data |  7qv6MC  7qv5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14168.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14168.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14168_msk_1.map emd_14168_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_14168_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14168_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14168_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : aurein 3.3
| Entire | Name: aurein 3.3 |
|---|---|
| Components |
|
-Supramolecule #1: aurein 3.3
| Supramolecule | Name: aurein 3.3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: Ranoidea raniformis (blue-thighed treefrog) / Synthetically produced: Yes |
| Molecular weight | Theoretical: 22.3 kDa/nm |
-Macromolecule #1: Aurein-3.3
| Macromolecule | Name: Aurein-3.3 / type: protein_or_peptide / ID: 1 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Ranoidea raniformis (blue-thighed treefrog) |
| Molecular weight | Theoretical: 1.800171 KDa |
| Sequence | String: GLFDIVKKIA GHIVSSI UniProtKB: Aurein-3.3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: wait time 15s after application, blot time 4s. |
| Details | Concentration refers to peptide monomer before fibrillation |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 15 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 5514 / Average exposure time: 2.0 sec. / Average electron dose: 40.0 e/Å2 / Details: Collected in movie-mode; 40 frames per exposure. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.1 µm / Calibrated defocus min: 0.35000000000000003 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | - Initial model building of a single helical layer using ChimeraX and Coot - Expansion to three layers, refinement in ISOLDE - Truncation of outer layers, re-expansion to three layers, refinement using phenix.real_space_refine |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 78.37 / Target criteria: CC |
| Output model |  PDB-7qv6: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)