+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | TIR-SAVED effector bound to cA3 | ||||||||||||
 Map data Map data | sharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Microbacterium ketosireducens TIR SAVED complex bound to cA3 / SIGNALING PROTEIN | ||||||||||||
| Function / homology | SMODS-associated and fused to various effectors / SMODS-associated and fused to various effectors sensor domain / TIR domain / Toll/interleukin-1 receptor homology (TIR) domain / Toll/interleukin-1 receptor homology (TIR) domain superfamily / signal transduction / TIR domain-containing protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Microbacterium ketosireducens (bacteria) Microbacterium ketosireducens (bacteria) | ||||||||||||
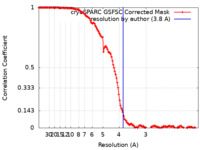
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||
 Authors Authors | Spagnolo L / White MF / Hogrel G | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Cyclic nucleotide-induced helical structure activates a TIR immune effector. Authors: Gaëlle Hogrel / Abbie Guild / Shirley Graham / Hannah Rickman / Sabine Grüschow / Quentin Bertrand / Laura Spagnolo / Malcolm F White /    Abstract: Cyclic nucleotide signalling is a key component of antiviral defence in all domains of life. Viral detection activates a nucleotide cyclase to generate a second messenger, resulting in activation of ...Cyclic nucleotide signalling is a key component of antiviral defence in all domains of life. Viral detection activates a nucleotide cyclase to generate a second messenger, resulting in activation of effector proteins. This is exemplified by the metazoan cGAS-STING innate immunity pathway, which originated in bacteria. These defence systems require a sensor domain to bind the cyclic nucleotide and are often coupled with an effector domain that, when activated, causes cell death by destroying essential biomolecules. One example is the Toll/interleukin-1 receptor (TIR) domain, which degrades the essential cofactor NAD when activated in response to infection in plants and bacteria or during programmed nerve cell death. Here we show that a bacterial antiviral defence system generates a cyclic tri-adenylate that binds to a TIR-SAVED effector, acting as the 'glue' to allow assembly of an extended superhelical solenoid structure. Adjacent TIR subunits interact to organize and complete a composite active site, allowing NAD degradation. Activation requires extended filament formation, both in vitro and in vivo. Our study highlights an example of large-scale molecular assembly controlled by cyclic nucleotides and reveals key details of the mechanism of TIR enzyme activation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14122.map.gz emd_14122.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14122-v30.xml emd-14122-v30.xml emd-14122.xml emd-14122.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14122_fsc.xml emd_14122_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14122.png emd_14122.png | 70.8 KB | ||
| Masks |  emd_14122_msk_1.map emd_14122_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14122.cif.gz emd-14122.cif.gz | 6.6 KB | ||
| Others |  emd_14122_additional_1.map.gz emd_14122_additional_1.map.gz emd_14122_additional_2.map.gz emd_14122_additional_2.map.gz emd_14122_half_map_1.map.gz emd_14122_half_map_1.map.gz emd_14122_half_map_2.map.gz emd_14122_half_map_2.map.gz | 4.8 MB 120.4 MB 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14122 http://ftp.pdbj.org/pub/emdb/structures/EMD-14122 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14122 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14122 | HTTPS FTP |
-Related structure data
| Related structure data |  7qqkMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14122.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14122.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.997 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14122_msk_1.map emd_14122_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: local filter map
| File | emd_14122_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local filter map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map
| File | emd_14122_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half mapB
| File | emd_14122_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half mapB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_14122_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : one tier of TIR_SAVED bound to cA3
| Entire | Name: one tier of TIR_SAVED bound to cA3 |
|---|---|
| Components |
|
-Supramolecule #1: one tier of TIR_SAVED bound to cA3
| Supramolecule | Name: one tier of TIR_SAVED bound to cA3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Microbacterium ketosireducens (bacteria) Microbacterium ketosireducens (bacteria) |
-Macromolecule #1: TIR_SAVED fusion protein
| Macromolecule | Name: TIR_SAVED fusion protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Microbacterium ketosireducens (bacteria) Microbacterium ketosireducens (bacteria) |
| Molecular weight | Theoretical: 46.815875 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPDTAINPRD PVFVSYRHSD GIALAAELTW LLRAAGIPVW RDVDDLPPGD TDARLQQAID EGISGAVIII TPQIADSRVV REVEAPRLL RLHRSSPQFA LGIVNAIQTS TGVVDYDAPD RVLGMERPEL RSVDQKSASR LGLVTMARQM LWHRIAAIRP L LSASGGEL ...String: MPDTAINPRD PVFVSYRHSD GIALAAELTW LLRAAGIPVW RDVDDLPPGD TDARLQQAID EGISGAVIII TPQIADSRVV REVEAPRLL RLHRSSPQFA LGIVNAIQTS TGVVDYDAPD RVLGMERPEL RSVDQKSASR LGLVTMARQM LWHRIAAIRP L LSASGGEL RLSLQTRNTP QVYDRTDADL DIRIRPSAHE KLPSAHGLED FAETAQFLPD AVTRAGANGV RIEGGAHLSV SI AIGAAIP STRVGPMTVV DGRGVHWVSS TEPQLPDEPR LRIVRESTIP STAPAPGRPD VAAYIDLQHP RSDAAFDNYL TEH AAELVA WQHLAPTRTG LLDAADGGTI AAEAVAHIRE LSMTNGNAVV HLMVRGPFGL AVLIGRLTNT LRVVAYEWTD SDAP DGTFM PPRYEPIVQL RASTPAGVIE RVIVADAE UniProtKB: TIR domain-containing protein |
-Macromolecule #2: RNA (5'-R(P*AP*AP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*AP*A)-3') / type: rna / ID: 2 / Number of copies: 4 |
|---|---|
| Source (natural) | Organism:  Microbacterium ketosireducens (bacteria) Microbacterium ketosireducens (bacteria) |
| Molecular weight | Theoretical: 942.66 Da |
| Sequence | String: AAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-64 (8k x 8k) / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.5 µm |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)