[English] 日本語
 Yorodumi
Yorodumi- EMDB-14022: Volta phase plate cryo-ET of Magnetospirillum gryphiswaldense ove... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14022 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Volta phase plate cryo-ET of Magnetospirillum gryphiswaldense overproducing PopZ-Mgr | |||||||||
 Map data Map data | Volta phase plate cryo-electron tomography of Magnetospirillum gryphiswaldense (wild-type) overproducing PopZ-Mgr | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PopZ / Alphaproteobacteria / cytoskeleton / polarity / CELL CYCLE | |||||||||
| Biological species |  Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Toro-Nahuelpan M / Plitzko JM / Schueler D / Pfeiffer D | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2022 Journal: J Mol Biol / Year: 2022Title: In vivo Architecture of the Polar Organizing Protein Z (PopZ) Meshwork in the Alphaproteobacteria Magnetospirillum gryphiswaldense and Caulobacter crescentus. Authors: Mauricio Toro-Nahuelpan / Jürgen M Plitzko / Dirk Schüler / Daniel Pfeiffer /  Abstract: The polar organizing protein Z (PopZ) forms a polar microdomain that is inaccessible to larger macromolecules such as ribosomes, and selectively sequesters proteins crucial for cell cycle control and ...The polar organizing protein Z (PopZ) forms a polar microdomain that is inaccessible to larger macromolecules such as ribosomes, and selectively sequesters proteins crucial for cell cycle control and polar morphogenesis in various Alphaproteobacteria. However, the in vivo architecture of this microdomain has remained elusive. Here, we analyzed the three-dimensional ultrastructural organization of the PopZ network in Magnetospirillum gryphiswaldense and Caulobacter crescentus by Volta phase plate cryo-electron tomography, which provides high spatial resolution and improved image contrast. Our results suggest that PopZ forms a porous network of disordered short, flexible, and branching filaments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14022.map.gz emd_14022.map.gz | 196.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14022-v30.xml emd-14022-v30.xml emd-14022.xml emd-14022.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14022.png emd_14022.png | 52 KB | ||
| Masks |  emd_14022_msk_1.map emd_14022_msk_1.map | 1 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-14022.cif.gz emd-14022.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14022 http://ftp.pdbj.org/pub/emdb/structures/EMD-14022 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14022 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14022 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10885 (Title: In vivo architecture of the polar organizing protein Z (PopZ) meshwork in the Alphaproteobacteria Magnetospirillum gryphiswaldense and Caulobacter crescentus EMPIAR-10885 (Title: In vivo architecture of the polar organizing protein Z (PopZ) meshwork in the Alphaproteobacteria Magnetospirillum gryphiswaldense and Caulobacter crescentusData size: 6.3 Data #1: Volta phase plate cryo-electron tomography of Magnetospirillum gryphiswaldense (wild-type) overproducing PopZ-Mgr [reconstructed volumes] Data #2: Volta phase plate cryo-electron tomography of Magnetospirillum gryphiswaldense delta-mamK overproducing PopZ-Mgr [reconstructed volumes] Data #3: Volta phase plate cryo-electron tomography of purified PopZ-Mgr [reconstructed volumes] Data #4: Cryo-electron tomography of Magnetospirillum gryphiswaldense delta-popZ overproducing PopZ-Mgr [reconstructed volumes] Data #5: Volta phase plate cryo-electron tomography of Caulobacter crescentus NA1000 [reconstructed volumes] Data #6: Volta phase plate cryo-electron tomography of Caulobacter crescentus NA1000 overproducing PopZ-Cc [reconstructed volumes] Data #7: Cryo-electron tomography of Magnetospirillum gryphiswaldense delta-popZ [reconstructed volumes] Data #8: Volta phase plate cryo-electron tomography of Magnetospirillum gryphiswaldense delta-mamK-mamY [reconstructed volumes]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14022.map.gz / Format: CCP4 / Size: 263.6 MB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_14022.map.gz / Format: CCP4 / Size: 263.6 MB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Volta phase plate cryo-electron tomography of Magnetospirillum gryphiswaldense (wild-type) overproducing PopZ-Mgr | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 13.68 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_14022_msk_1.map emd_14022_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
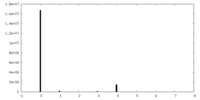
| Projections & Slices |
| ||||||||||||
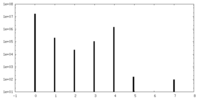
| Density Histograms |
- Sample components
Sample components
-Entire : Magnetospirillum gryphiswaldense (wild-type) overproducing PopZ-Mgr
| Entire | Name: Magnetospirillum gryphiswaldense (wild-type) overproducing PopZ-Mgr |
|---|---|
| Components |
|
-Supramolecule #1: Magnetospirillum gryphiswaldense (wild-type) overproducing PopZ-Mgr
| Supramolecule | Name: Magnetospirillum gryphiswaldense (wild-type) overproducing PopZ-Mgr type: cell / ID: 1 / Parent: 0 Details: Individual segmented structures are depicted using different colors in the EMDB entry page image and in the corresponding publication: Putative PopZ filaments are depicted in white. MamK ...Details: Individual segmented structures are depicted using different colors in the EMDB entry page image and in the corresponding publication: Putative PopZ filaments are depicted in white. MamK filaments are green. The flagellum is colored in gold. The cellular envelope inner and outer membranes are depicted in blue. |
|---|---|
| Source (natural) | Organism:  Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) |
-Supramolecule #2: PopZ
| Supramolecule | Name: PopZ / type: organelle_or_cellular_component / ID: 2 / Parent: 1 Details: The polar organizing protein Z (PopZ) forms a polar microdomain that is inaccessible to larger macromolecules such as ribosomes, and selectively sequesters proteins crucial for cell cycle ...Details: The polar organizing protein Z (PopZ) forms a polar microdomain that is inaccessible to larger macromolecules such as ribosomes, and selectively sequesters proteins crucial for cell cycle control and polar morphogenesis in various Alphaproteobacteria. In the present strain PopZ overproduction was achieved via insertion of a Tn5-Ptet-based popZ overexpression cassette into the genome of the Magnetospirillum gryphiswaldense wild-type strain. |
|---|---|
| Source (natural) | Organism:  Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) |
-Supramolecule #3: MamK
| Supramolecule | Name: MamK / type: organelle_or_cellular_component / ID: 3 / Parent: 1 Details: The filament-forming bacterial actin MamK is important for organizing magnetosome organelles into chains that are used for navigation along geomagnetic field lines. Magnetosomes are ...Details: The filament-forming bacterial actin MamK is important for organizing magnetosome organelles into chains that are used for navigation along geomagnetic field lines. Magnetosomes are membranous organelles containing nanometer-sized crystals of magnetite present in magnetotactic bacteria. |
|---|---|
| Source (natural) | Organism:  Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) |
-Supramolecule #4: Cellular envelope
| Supramolecule | Name: Cellular envelope / type: organelle_or_cellular_component / ID: 4 / Parent: 1 / Details: inner and outer membranes |
|---|---|
| Source (natural) | Organism:  Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) |
-Supramolecule #5: Flagellum
| Supramolecule | Name: Flagellum / type: organelle_or_cellular_component / ID: 5 / Parent: 1 Details: Flagella are helical protein filaments powered by a rotary motor to mediate motility of bacteria. |
|---|---|
| Source (natural) | Organism:  Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) Magnetospirillum gryphiswaldense MSR-1 (magnetotactic) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: modified flask standard medium (FSM) |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: MOLYBDENUM / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE Details: The mixture was blotted and embedded in vitreous ice by plunge freezing into liquid ethane (< - 170 C). The grids were stored in sealed boxes in liquid nitrogen until used.. |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: Sigma-Aldrich / Diameter: 15 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Tomography was performed under low-dose conditions using a Titan Krios transmission electron microscope (FEI) equipped with a 300 kV field emission gun, and a Gatan Quantum post-column energy filter. Tilt series were acquired using Serial EM software. The specimen was tilted about one axis with 1.5 degree increments over a typical total angular range of -+ 60 degree. To account for the increased specimen thickness at high tilt angles, the exposure time was multiplied by a factor of 1/cos alpha. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.5 e/Å2 Details: Data collection was performed at 300 kV, with the energy filter operated in the zero-loss mode (slit width of 20 eV). The cumulative electron dose during the tilt series was kept below 150 e- A-2. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Tomograms were reconstructed with (15 nm gold fiducials) the IMOD software package (https://bio3d.colorado.edu/imod/) and treated with an anisotropic non-linear diffusion denoising algorithm to improve signal-to-noise ratio (K: 1, Iterations: 10). Segmentation was performed using Amira software on binned volumes with a voxel size of 10.48 A and 13.68 A. Filaments were traced in Amira using an automated segmentation algorithm based on a generic cylinder as a template implemented in the X-Tracing extension. Prior to filament tracing, binned volumes were subjected to nonlocal-means filtering using Amira software (Thermo Fisher Scientific). The cylindrical templates were generated with a diameter and length of 6 and 15 nm, respectively. To reduce background noise, short filamentous structures with lengths below 30 nm were filtered out. Membrane segmentation was done using the software TomoSegMemTV and a complementary package, SynapSegTools, both for Matlab, and refined manually in Amira (Thermo Fisher Scientific). |
|---|---|
| Final reconstruction | Number images used: 321 |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X